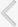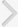|
 |


- Search
| Int Neurourol J > Volume 20(Suppl 1); 2016 > Article |
|
ABSTRACT
Purpose:
We aim to fabricate a thermoplastic poly(methylmethacrylate) (PMMA) Lab-on-a-Chip device to perform continuous- flow polymerase chain reactions (PCRs) for rapid molecular detection of foodborne pathogen bacteria.
Methods:
A miniaturized plastic device was fabricated by utilizing PMMA substrates mediated by poly(dimethylsiloxane) interfacial coating, enabling bonding under mild conditions, and thus avoiding the deformation or collapse of microchannels. Surface characterizations were carried out and bond strength was measured. The feasibility of the Lab-on-a-Chip device for performing on-chip PCR utilizing a lab-made, portable dual heater was evaluated. The results were compared with those obtained using a commercially available thermal cycler.
Results:
A PMMA Lab-on-a-Chip device was designed and fabricated for conducting PCR using foodborne pathogens as sample targets. A robust bond was established between the PMMA substrates, which is essential for performing miniaturized PCR on plastic. The feasibility of on-chip PCR was evaluated using Escherichia coli O157:H7 and Cronobacter condimenti, two worldwide foodborne pathogens, and the target amplicons were successfully amplified within 25 minutes.
Conclusions:
In this study, we present a novel design of a low-cost and high-throughput thermoplastic PMMA Lab-on-a-Chip device for conducting microscale PCR, and we enable rapid molecular diagnoses of two important foodborne pathogens in minute resolution using this device. In this regard, the introduced highly portable system design has the potential to enable PCR investigations of many diseases quickly and accurately.
Infectious diseases continue to place a significant global health and economic burden on society. These concerns provoke the need for developing effective and reliable methods to quickly diagnose different diseases. For example, diagnosis of schistosomiasis is normally based on the detection of the presence of parasite eggs via microscopic examinations of stool or urine samples. Serologic tests can often help improve the outcome of this technique. However, such assays exhibit certain drawbacks, including low sensitivities and difficulties in sample collection for analysis, which potentially hampers the efficiency of such assays [1]. Another example is the diagnosis of prostate cancer, which presents notable challenges even though prostate cancer is frequently diagnosed by direct rectal examination. Early prostate cancers usually do not cause symptoms, but more progressive cancers are sometimes first found because of the symptoms they cause. There is a screening test that looks for the level of the prostate-specific antigen in serum. But this test has only an 85% predictive value and men who many have high levels may not progress to having cancer [2].
Owing to polymerase chain reaction (PCR) [3-6], simple yet sensitive diagnostic tools have been developed to target specific genes responsible for different infectious diseases. Furthermore, recent advances in Micro Total Analysis Systems possess a number of advantages including the use of small reagent volumes, rapid analysis, small footprint of the device, low power consumption, parallel analyses, and functional integration of multiple devices [7-10]. As one major application, foodborne pathogens have been widely detected using Lab-on-a-Chip devices. Escherichia coli O157:H7 is a foodborne pathogen of major significance that is attributed to the production of Shiga toxins [11,12] which leads to illness, typically through the consumption of contaminated and raw food including raw leaf green vegetables, undercooked meat and raw milk. Infections may lead to hemorrhagic diarrhea, and to kidney failure. Cronobacter is a group of opportunistic pathogens that causes infections in all age groups but is more common in adults than in infants [13]. But higher mortality is seen in infants. The demand for performing simple, inexpensive, and rapid detection of pathogens such as these has led to the widespread application of PCR because of its high specificity, sensitivity, and efficiency, particularly its ability to successfully detect low amounts of target DNA.
In this study, we fabricated a poly(methylmethacrylate) (PMMA) Lab-on-a-Chip device to perform continuous-flow PCR for rapid detection of foodborne pathogens. PMMA was used as an alternative material to replace glasses for a wide range of applications, such as biomedical applications, optical applications, and sensors [14], and it has been used for fabricating microfluidic chips, especially for PCR analysis [15-18]. Despite its many advantages, PMMA has some limitations in bonding. Among many bonding methods used for PMMA [19-23], thermal bonding techniques have been widely employed mainly due to simplicity in the process. However, the method requires heating the substrate above the glass transition temperature (Tg, 105˚C) [24,25] under a high-pressure condition [20]. Thus, the microchannels can be slightly deformed. To overcome this problem, we adopted an intermediate layer containing of a poly(dimethylsiloxane) (PDMS) structure, as demonstrated in our previous study [26]. In short, the PMMA substrates were first treated with aminosilane, a silane coupling reagent containing amine functionality, and subsequently reacted with low-molecular-weight PDMS (epoxy-PDMS) to graft PDMS functionality onto the surface of the APTES-coated PMMA substrates. During this process, the inner surface of the microchannel becomes similar to PDMS, which possesses the advantages of suppressing bubbles generated during the PCR process. To examine the feasibility of the Lab-on-a-Chip device in a “proof of concept manner,” a two-temperature PCR assay [18,27] was performed using a lab-made dual temperature heater using E. coli O157:H7 and Cronobacter condimenti.
PMMA sheets (2 mm thick) were purchased from Goodfellow (Huntingdon, England). PDMS prepolymer (Sylgard 184) and a curing agent were purchased from Dow Corning (Midland, MI, USA). 3-Aminopropyltriethoxysilane (APTES, 99%), monoglycidyl ether terminated PDMS (average Mn 5,000 g/mol), and bovine serum albumin (BSA; V fraction) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Taq polymerase, PCR buffer solutions, deoxynucleotides (dNTPs), and genomic DNA purification kit were purchased from Promega (Madison, WI, USA). Luria-Bertani (LB) broth was purchased from MB cell (Los Angeles, CA, USA), and trypticase soy broth was purchased from Becton Dickinson (Franklin Lakes, NJ, USA). A 100-bp DNA size marker was purchased from Takara (Shiga, Japan). Agarose powder was purchased from BioShop (Burlington, ON, Canada). Ethidium bromide dye (Loading STAR) used for staining DNA was purchased from Dynebio (Seongnam, Korea). Robotic QIAcube workstation was purchased from Qiagen (Germantown, MD, USA).
A serpentine microchannel was engraved on a PMMA substrate (35 mm×45 mm) using a computer numerical control milling machine. The width, depth, and total length of the microchannel were designed to be 300 μm, 100 μm, and 180 cm, respectively, comprising 30 thermal cycles. Silicone tubes (inner diameter, 1 mm; outer diameter, 2 mm) were inserted into the inlet and outlet ports. In the experiment, a two-temperature PCR assay was performed using a lab-made portable heater.
Fig. 1 schematically shows the procedures for hydrophobizing PMMA surfaces and bonding two PMMA substrates via PDMS interfacial coating. The PMMA substrates were cleaned by sonication in water for 5 minutes. They were then treated with oxygen plasma for 2 minutes and dipped in a 5% (v/v) aqueous solution of APTES at 80°C for 20 minutes, as shown in Fig. 1A. After the surface was cleaned with a copious amount of distilled water and dried completely by compressed air, the APTES-coated PMMA substrates were reacted with the epoxy-PDMS at 80°C for 4 hours to form a robust amine-epoxy bond (Fig. 1B). The unreacted PDMS was removed by sonication in isopropyl alcohol for 1 minute. Subsequently, as shown in Fig. 1C and D, two PDMS-coated PMMA substrates were oxidized and brought into conformal contact with each other at 80°C for 20 minutes under a pressure of 0.1 MPa.
The water contact angles were measured on the surfaces of pristine PDMS and PMMA substrates, plasma-treated, APTES-coated, and epoxy-PDMS-coated PMMA substrates by the sessile drop technique using a Phoenix 300 contact angle measuring system (Surface Electro Optics, Suwon, Korea). The measurements were performed in triplicate.
The modified PMMA surfaces were analyzed by fourier transform infrared spectroscopy (FTIR; JASCO 4700 FTIR spectrophotometer) which offers quantitative and qualitative analyses to characterize the chemical nature of modified surfaces. The FTIR indicated He-Ne as the reference at a resolution of 4/cm and the mode of diffuse reflectance with scanning from 4,500 to 400/cm.
To evaluate the bond strength of the PMMA substrates (20 mm×20 mm), a peel test was conducted using a texture analyzer (QTS 25, Brookfield, Middleboro, MA, USA). The bonded assembly was pulled at a speed of 100 mm/min. All measurements were performed in triplicate. A leakage test was conducted by injecting an ink solution into the PMMA microchannel using a syringe pump (KDS 200, KD Scientific, New Hope, PA, USA). The flow rates were systematically controlled at 0.2, 2.0, 20, and 40 mL/min.
For application in PCR processes, purified C. condimenti and E. coli O157:H7 DNA solutions were used as DNA templates. Briefly, Cronobacter strains were grown overnight at 37°C in 5 mL of trypticase soy broth supplemented with 1% NaCl, shaking at 160 rpm. Genomic DNA was isolated from 2 mL of the culture using a robotic QIAcube workstation with its automated Qiagen DNeasy chemistry following the manufacturer’s recommendations. Typically, 5–15 ng/μL of purified genomic DNA was recovered in a final elution volume of 200 μL. E. coli O157:H7 were grown overnight at 37°C in 5 mL of LB broth with low salt, shaking at 200 rpm, and the genomic DNA of E. coli O157:H7 was extracted from 2 mL of the culture using genomic DNA purification kit. Input DNA with concentrations of 3 ng/μL (C. condimenti) and 1 ng/μL (E. coli O157:H7) were added to PCR reaction mixture. The primer sequences for amplifying a 210 bp DNA fragment of Shiga-toxin gene in E. coli O157:H7 were as follows: 5ʹ-TGT AAC TGG AAA GGT GGA GTA TAC A–3ʹ (forward) and 5ʹ-GCT ATT CTG AGT CAA CGA AAA ATA AC–3ʹ (reverse). The primer sequences for amplifying a 689-bp gene fragment of rpoB gene in the C. condimenti were as follows: 5ʹ-AAC GCC AAG CCA ATC TCG–3ʹ (forward) and 5ʹ-GTA CCG CCA CGT TTT GCT–3ʹ (reverse) [28]. The PCR mixture contained 5-μL buffer, 0.16mM dNTPs mixture, 0.5 μM forward and reverse primers, and 0.5 U/μL of Taq polymerase. Amplifications were performed for 30 thermal cycles. The inner surface of the microchannel was treated with a BSA solution (1.5 mg/mL) to reduce the nonspecific adsorption of the PCR reagent on the walls of the microchannel. Then, the PCR reagent was introduced into the microchannel at a flow rate of 2 µL/min. PCR amplicons were subjected to agarose gels in a RunOne (Embi Tec, San Diego, CA, USA) horizontal electrophoresis unit and were photographed with trans-illuminated UV light using a Bio-Rad Molecular Imager Gel-Chem-Doc XR imaging system (Bio-Rad Laboratories, Hercules, CA, USA).
Fig. 2 shows the results of the water contact angle measurements performed on the surfaces of PDMS (pristine) and PMMA (pristine, oxygen plasma-treated, APTES-coated, and epoxy-PDMS-coated) substrates. PMMA was used as the model substrate and pristine PDMS as a reference. The average water contact angles measured on the surfaces of the pristine PDMS and PMMA were 99.8˚±3.4˚ (Fig. 2A) and 73.2˚±2.1˚ (Fig. 2B), respectively. The water contact angles for PMMA decreased to 43.6˚±2.4˚ after plasma treatment for 1 minute and increased to 68.7˚±3.2˚ after treatment with 5% APTES. Based on these results, we could confirm that the amine functionalities were successfully anchored onto the PMMA surface [26]. After the PMMA substrate was treated with a monolayer of PDMS, the water contact angles of PMMA increased to 94.9˚±2.0˚, which was almost equivalent to that of the pristine PDMS, 99.8˚±3.4˚, as shown in Fig. 2A. These results indicate that a PDMS monolayer was successfully generated on the PMMA surface.
Fig. 3 shows the results of FTIR analyses performed on both pristine and modified PMMA, as well as on pristine PDMS. The assignments of the bands are shown in Tables 1 and 2. Fig. 3 depicts the FTIR spectroscopic analysis of pristine PMMA (Fig. 3A), APTES-coated PMMA (Fig. 3B), epoxy-PDMS-coated PMMA (Fig. 3C), and pristine PDMS (Fig. 3D). Epoxy-PDMS-coated PMMA is defined throughout the text as the substrate coated with an epoxy-PDMS layer on an APTES-coated PMMA. Fig. 3E compares the peaks for pristine PMMA, APTES-coated PMMA, and epoxy-PDMS-coated PMMA. Fig. 3F compares the peaks for pristine PDMS and epoxy-PDMS-coated PMMA.
Pristine PMMA (Fig. 3A) exhibited spectral regions around 1,080–1,202, 1,232–1,274, and 1,725/cm, which corresponded to the presence of C–O–C, C–O, and C=O stretching vibrations of the ester group, respectively [29-31]. Absorption bands corresponding to the –CH2 twisting modes of PMMA appeared around 760–960/cm. In addition, the spectral peak at 2,948/cm confirmed the formation of the primary C−H stretching vibration [32,33]. The FTIR spectrum of the APTES-coated PMMA (Fig. 3B) displayed a new peak at 1,642/cm, indicating the – NH2 scissors bending vibration attributed to the 3-APTES coating [32,33]. The position of the typical bands at 647–1,074/cm indicated the presence of Si–O–Si stretching modes on the APTES-coated PMMA [29-31]. In total, these results confirm that self-assembled layers containing silane radicals and amino radicals were successfully formed on the PMMA surface.
Infrared (IR) spectra after the coating with epoxy-PDMS (Fig. 3C) were obtained in accordance with the bonding mechanism anticipated to take place between PDMS and APTES-coated PMMA. Strong absorption broadband regions at 690–826 and 1,049–1,005/cm were associated with the presence of −Si−C− and the Si−O−Si stretching vibration, respectively, thus confirming the crosslinking between the amine and epoxy functionalities [34]. Moreover, the peaks corresponding to the wavelengths of 1,259, 1,406, and 2,960/cm were assigned to the symmetric –CH3 deformation in –Si–CH3, the asymmetric Si–CH3 bending, and the asymmetric –CH3 stretching, respectively [34,35]. The amide –NH2 scissors bending vibration at 1,642/cm was not identified on any of the surfaces analyzed. The same absorption bands were exhibited for epoxy-PDMS-coated PMMA (Fig. 3C) and pristine PDMS (Fig. 3D), confirming that surface modification was successfully realized on PMMA.
Fig. 4 shows the results of bond strength analyses. Fig. 4A shows the experimental setup for performing the peel test. The inset in Fig. 4A shows bonded PMMAs. Fig. 4B shows the photo taken at the moment of substrate detachment. The average bond strength was calculated to be approximately 214.4 kPa. The red rectangle in Fig. 4C shows the area previously bonded before the detachment. The area was approximately 308 mm2. Fig. 4D shows the black ink solution injected into the microchannel, and Fig. 4E shows the overall setup for performing the leakage test. The flow rates of the ink injection was varied at 0.2, 2.0, 20, and 40 mL/min, corresponding to per minute injection volumes of 10, 100, 1,000, and 2,000 times the total internal volume of the microchannel used in the leakage test. No leak was observed even at the highest flow rate examined, demonstrating the adaptive potential that this device possesses for applications where high speed sample injections are desirable.
A lab-made dual temperature heater was used for heating the Lab-on-a-Chip device, as shown in Fig. 5A. Two isolated aluminum blocks were heated by two 30-W positive-temperature-coefficient (PTC) heaters (DBK HP05-1/09-240) (Fig. 5B). The heat blocks were maintained at temperatures of 95˚C and 60˚C, and controlled by turning on and off the individual PTC heaters using a proportional-integral-derivative control device. The temperatures were measured by one-wire digital thermometers (DS18S20) located in each heat block. To evaluate the precision of the temperature, a PMMA substrate (4 cm in width, 8 cm in length, and 0.2 cm in thickness) was used as a model substrate for the measurement of temperature (Fig. 5C). The average temperatures measured in the sectioned areas of the denaturation and annealing/extension were 94.6°C ±0.5°C and 60.1°C ±1.4°C. The corresponding coefficients of variation (CVs) for the temperature measurement precision were 0.54% (n =20) and 2.3% (n=20).
The images in Fig. 6 show the bubble suppression phenomena under actual heated conditions with the flow of red ink inside the PDMS-PDMS Lab-on-a-Chip device, PMMA-PMMA Lab-on-a-Chip device, and PMMA-PMMA Lab-on-a-Chip device coated with epoxy-PDMS onto the inner walls of the microchannels when 5, 15, and 30 minutes have elapsed after the introduction of the red ink, respectively. As shown in Fig. 6, the PDMS microchannel produced a lot of bubbles, disconnecting the sample plug from place to place owing to the high gas permeability of the PDMS material itself. On the other hand, the PMMA microchannels with and without coating with epoxy-PDMS better prevented formation of air bubbles as compared to the PDMS microchannel due to the low gas permeability properties of the PMMA material itself. Bubble formation was completely suppressed inside the PMMA microchannel coated with epoxy-PDMS. These results indicated that the coating of the PMMA microchannel with epoxy-PDMS aided in maintaining the homogeneous flow of liquid inside a microchannel during PCR.
Fig. 7A shows an illustration of the serpentine microchannel used for performing continuous-flow PCR. Fig. 7B shows a photo of the PMMA Lab-on-a-Chip device used in this experiment. A total of 20 μL of PCR reaction mixture was introduced into the microchannel and collected at the outlet port. Fig. 7C shows the overall system for performing continuous-flow PCR analysis using the PMMA Lab-on-a-Chip device. A lab-made heater was used for thermal cycling parameters for PCR, and a syringe pump was used for sample introduction into the Lab-on-a-Chip device. Fig. 7D shows an IR camera image of the Lab-on-a-Chip device illustrating the temperature profile simulated under actual PCR condition. In this study, two DNA targets (210 and 689 bp) were amplified under the same annealing/extension temperature owing to a similar Tm value (60˚C) of the primer pairs used for amplification of the two molecular targets. The average temperatures measured at the sectioned areas of the denaturation and annealing/extension zones were 94.3˚C±0.4˚C and 60.2˚C ±1.1˚C, respectively, and the corresponding CVs were 0.37% (n=15) and 1.8% (n=15). Fig. 7E shows the 210-bp gene fragments amplified from the Shiga-toxin gene in the E. coli O157:H7. The total reaction took approximately 25 minutes. Although the average intensity of the target amplicon obtained using the PMMA Lab-on-a-Chip device was approximately 54.9% of that obtained using a thermal cycler, the target was still distinguishable without displaying non-specific bands or primer-dimers. Fig. 7F shows the 689-bp gene fragment amplified from rpoB gene in the C. condimenti strain. The average intensity of the target amplicon obtained using the PMMA Lab-on-a-Chip device was approximately 99.9% of that obtained using a thermal cycler. Based on these results, the fabricated PMMA Lab-on-a-Chip device with a modified inner microchannel mimicking the characteristics of PDMS reliably could amplify the two DNA targets with near identical intensities compared with that obtained using a thermal cycler. In both Fig. 7E and F, lane M shows 100-bp DNA size marker, and lane 1 shows the results of negative control (no template added) reactions.
A traditional molecular diagnostic technique, PCR, was successfully transferred onto a miniaturized platform constructed from inexpensive and mass-producible plastic under relatively mild temperature and pressure conditions assisted by surface modification of PMMA. The plastic Lab-on-a-Chip device was adopted as a portable diagnostic platform for the rapid identification of foodborne pathogens as a “proof-of-concept” experiment. Furthermore, a miniaturized and portable PCR device possesses considerable potential as an inexpensive and clean device free of sample contamination, not restricted to analyses of pathogens, but also in many clinical areas where immediate responses are demanded, including diagnosis of various types of infectious diseases.
NOTES
Grant Support
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2014R1A1A3051319) and the Public Welfare & Safety Research program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2012M3A2A1051681).
REFERENCES
1. Sandoval N, Siles-Lucas M, Perez-Arellano JL, Carranza C, Puente S, Lopez-Aban J, et al. A new PCR-based approach for the specific amplification of DNA from different Schistosoma species applicable to human urine samples. Parasitology 2006;133(Pt 5):581-7. PMID: 16834820


2. Fan MQ, Wang PX, Feng JY, Xiao Y, Huang CB. Biochip analysis of prostate cancer. Genet Mol Res 2014;13:152-9. PMID: 24446298


3. Smyth RP, Schlub TE, Grimm A, Venturi V, Chopra A, Mallal S, et al. Reducing chimera formation during PCR amplification to ensure accurate genotyping. Gene 2010;469:45-51. PMID: 20833233


4. Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 1986;51 Pt 1:263-73. PMID: 3472723


5. Sucher NJ, Deitcher DL. PCR and patch-clamp analysis of single neurons. Neuron 1995;14:1095-100. PMID: 7541630


6. Belinsky GS, Rich MT, Sirois CL, Short SM, Pedrosa E, Lachman HM, et al. Patch-clamp recordings and calcium imaging followed by single-cell PCR reveal the developmental profile of 13 genes in iPSC-derived human neurons. Stem Cell Res 2014;12:101-18. PMID: 24157591



7. Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, et al. Microfluidic diagnostic technologies for global public health. Nature 2006;442:412-8. PMID: 16871209


8. Chin CD, Linder V, Sia SK. Lab-on-a-chip devices for global health: past studies and future opportunities. Lab Chip 2007;7:41-57. PMID: 17180204


9. Tudos AJ, Besselink GJ, Schasfoort RB. Trends in miniaturized total analysis systems for point-of-care testing in clinical chemistry. Lab Chip 2001;1:83-95. PMID: 15100865


10. Belgrader P, Young S, Yuan B, Primeau M, Christel LA, Pourahmadi F, et al. A battery-powered notebook thermal cycler for rapid multiplex real-time PCR analysis. Anal Chem 2001;73:286-9. PMID: 11199979


11. Dharmasiri U, Witek MA, Adams AA, Osiri JK, Hupert ML, Bianchi TS, et al. Enrichment and detection of Escherichia coli O157:H7 from water samples using an antibody modified microfluidic chip. Anal Chem 2010;82:2844-9. PMID: 20218574



12. Abolmaaty A, Chen H, Faghri M. Microfluidic chip for direct detection of E.coli O157:H7 in ground beef via anti-digoxigenin immuno-PCR assay. World Appl Sci J 2011;14:591-8.
13. Yan Q, Jarvis KG, Chase HR, Hebert K, Trach LH, Lee C, et al. A proposed harmonized LPS molecular-subtyping scheme for Cronobacter species. Food Microbiol 2015;50:38-43. PMID: 25998813


14. Ali U, Karim KJ, Buang NA. A review of the properties and applications of poly (methyl methacrylate) (PMMA). Polym Rev 2015;55:678-705. 
15. Chen Y, Zhang L, Chen G. Fabrication, modification, and application of poly(methyl methacrylate) microfluidic chips. Electrophoresis 2008;29:1801-14. PMID: 18384069


16. Lounsbury JA, Karlsson A, Miranian DC, Cronk SM, Nelson DA, Li J, et al. From sample to PCR product in under 45 minutes: a polymeric integrated microdevice for clinical and forensic DNA analysis. Lab Chip 2013;13:1384-93. PMID: 23389252


17. Lounsbury JA, Landers JP. Ultrafast amplification of DNA on plastic microdevices for forensic short tandem repeat analysis. J Forensic Sci 2013;58:866-74. PMID: 23692541


18. Tran HH, Wu W, Lee NY. Ethanol and UV-assisted instantaneous bonding of PMMA assemblies and tuning in bonding reversibility. Sens Actuator B: Chem 2013;181:955-62. 
19. Chen Z, Gao Y, Lin J, Su R, Xie Y. Vacuum-assisted thermal bonding of plastic capillary electrophoresis microchip imprinted with stainless steel template. J Chromatogr A 2004;1038:239-45. PMID: 15233539


20. Kelly RT, Woolley AT. Thermal bonding of polymeric capillary electrophoresis microdevices in water. Anal Chem 2003;75:1941-5. PMID: 12713054


21. Kelly RT, Pan T, Woolley AT. Phase-changing sacrificial materials for solvent bonding of high-performance polymeric capillary electrophoresis microchips. Anal Chem 2005;77:3536-41. PMID: 15924386


22. Chen G, Li J, Qu S, Chen D, Yang P. Low temperature bonding of poly(methylmethacrylate) electrophoresis microchips by in situ polymerisation. J Chromatogr A 2005;1094:138-47. PMID: 16257300


23. Yussuf AA, Sbarski I, Hayes JP, Solomon M, Tran N. Microwave welding of polymeric-microfluidic devices. J Micromech Microeng 2005;15:1692-9. 
24. Martynova L, Locascio LE, Gaitan M, Kramer GW, Christensen RG, MacCrehan WA. Fabrication of plastic microfluid channels by imprinting methods. Anal Chem 1997;69:4783-9. PMID: 9406529


25. Galloway M, Stryjewski W, Henry A, Ford SM, Llopis S, McCarley RL, et al. Contact conductivity detection in poly(methyl methacrylate) -based microfluidic devices for analysis of mono- and polyanionic molecules. Anal Chem 2002;74:2407-15. PMID: 12038768


26. Zhang H, Lee NY. Non-silicon substrate bonding mediated by poly(dimethylsiloxane) interfacial coating. Appl Surf Sci 2015;327:233-40. 
27. Trinh KT, Wu W, Lee NY. Planar poly(dimethylsiloxane) (PDMS)-glass hybrid microdevice for a flow-through polymerase chain reaction (PCR) employing a single heater assisted by an intermediate metal alloy layer for temperature gradient formation. Sens Actuator B: Chem 2014;190:177-84. 
28. Lehner A, Fricker-Feer C, Stephan R. Identification of the recently described Cronobacter condimenti by an rpoB-gene-based PCR system. J Med Microbiol 2012;61(Pt 7):1034-5. PMID: 22466029


29. Riau AK, Mondal D, Yam GH, Setiawan M, Liedberg B, Venkatraman SS, et al. Surface modification of PMMA to improve adhesion to corneal substitutes in a synthetic core-skirt keratoprosthesis. ACS Appl Mater Interfaces 2015;7:21690-702. PMID: 26389670


30. Balamurugan A, Kannan S, Selvaraj V, Rajeswari S. Development and spectral characterization of poly(methyl methacrylate)/hydroxyapatite composite for biomedical applications. Trends Biomater Artif Organ 2004;18:41-5.
31. Majoul N, Aouida S, Bessaïs B. Progress of porous silicon APTES-functionalization by FTIR investigations. Appl Surf Sci 2015;331:388-91. 
32. Hsiao VK, Waldeisen JR, Zheng Y, Lloyd PF, Bunning TJ, Huang TJ. Aminopropyltriethoxysilane (APTES)-functionalized nanoporous polymeric gratings: fabrication and application in biosensing. J Mater Chem 2007;17:4896-901. 
33. Olmos D, Baselga J, Mondragon I, Gonzalez-Benito J. The effect of surface modification of silica microfillers in an epoxy matrix on the thermo-mechanical properties. J Adhes Sci Technol 2008;22:1443-59. 
34. Johnson LM, Gao L, Shields IV CW, Smith M, Efimenko K, Cushing K, et al. Elastomeric microparticles for acoustic mediated bioseparations. J Nanobiotechnology 2013;11:22. PMID: 23809852



35. Cho LL, Huang KB. Identification of condom lubricants by FT-IR Spectroscopy. Forensic Sci J 2012;11:33-40.
Fig. 1.
Schematic showing the bonding mechanism of two PMMA substrates through a PDMS interfacial coating. (A) Coating of APTES on PMMA. (B) Subsequent coating of epoxy-PDMS on APTES-coated PMMA. APTES and epoxy-PDMS were crosslinked via amine-epoxy bond formation. (C) Surface oxidation and subsequent contact of two epoxy-PDMS-coated PMMA substrates. (D) Bonding of two epoxy-PDMS-coated PMMA substrates by embossing at 80°C for 20 minutes under 0.1 MPa via siloxane bond (Si–O–Si) formation. PMMA, poly(methylmethacrylate); PDMS, poly(dimethylsiloxane); APTES, 3-aminopropyltriethoxysilane.
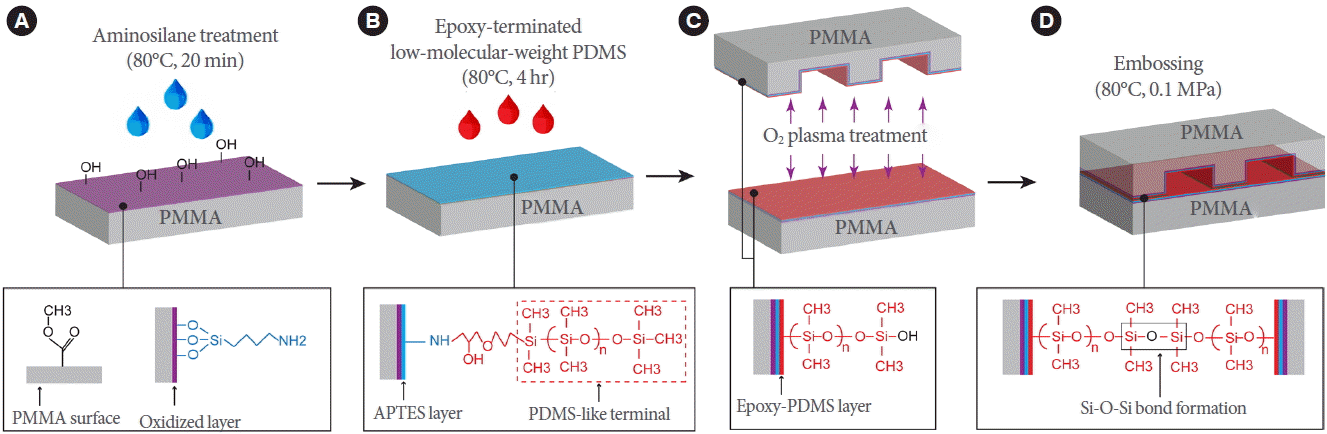
Fig. 2.
(A) Water contact angle measured on the surface of the pristine PDMS. (B) Water contact angles measured on the surfaces of the pristine, plasma-treated, APTES-coated, and epoxyPDMS-coated PMMA substrates. PMMA, poly (methylmethacrylate); PDMS, poly (dimethylsiloxane); APTES, 3-aminopropyltriethoxysilane.
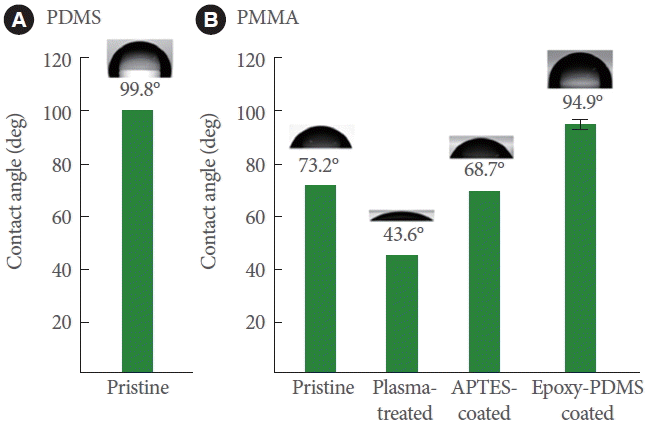
Fig. 3.
Fourier transform infrared spectroscopy spectra: (A) pristine PMMA, (B) APTES-coated PMMA, (C) epoxy-PDMS-coated PMMA, (D) pristine PDMS, (E) comparison among pristine PMMA, APTES-coated PMMA, and epoxy-PDMS-coated PMMA, (F) comparison between pristine PDMS and epoxy-PDMS-coated PMMA. PMMA, poly(methylmethacrylate); APTES, 3-aminopropyltriethoxysilane; PDMS, poly(dimethylsiloxane).
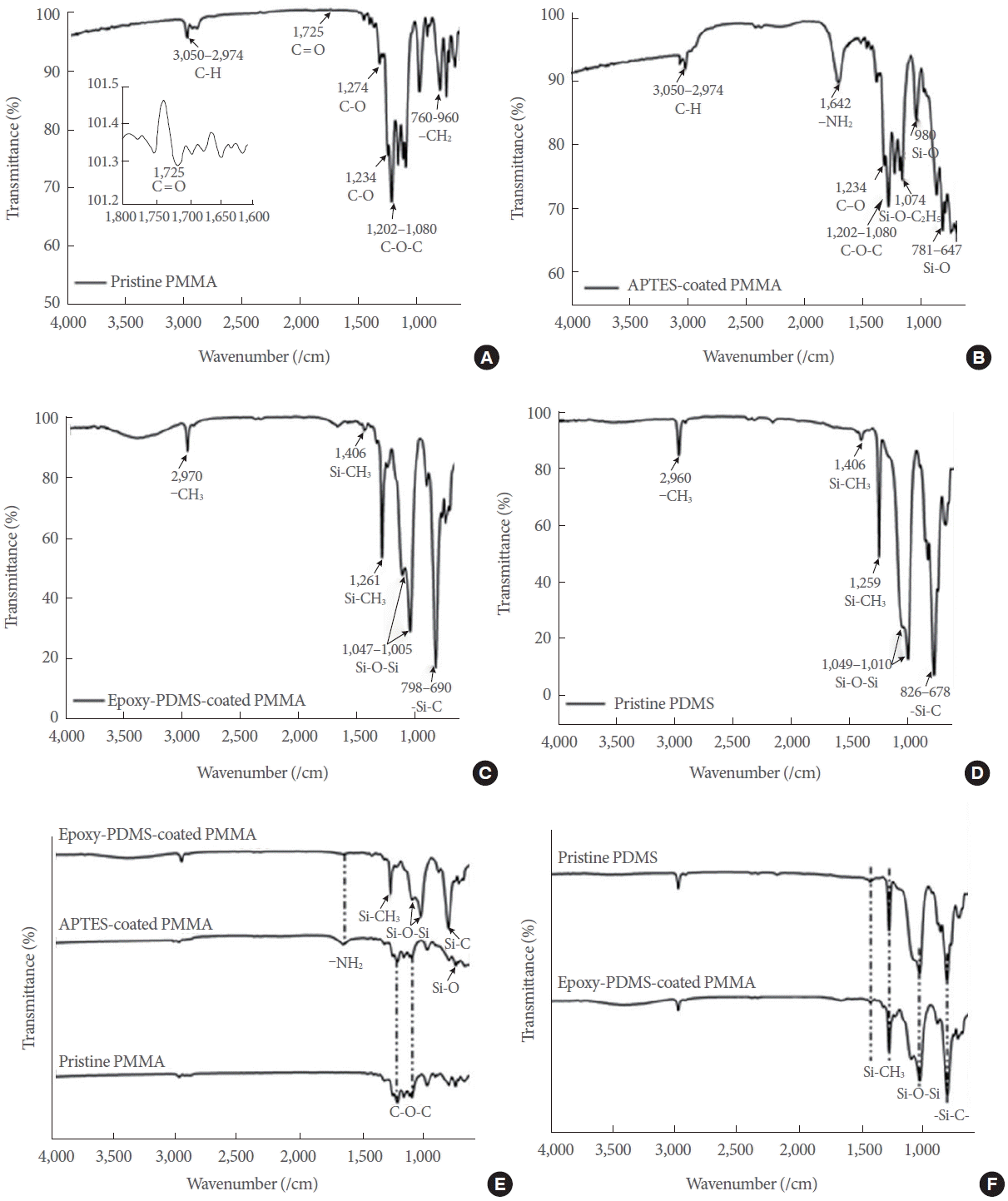
Fig. 4.
(A) Experimental setup for performing the peel test. (B) Photo of the substrates at the moment of detachment. (C) Photo of two poly(methylmethacrylate) (PMMA) after detachment (red rectangle shows bonded region). (D) Photo showing black ink injection into the microchannel. (E) Photo showing the overall setup for performing leakage test.
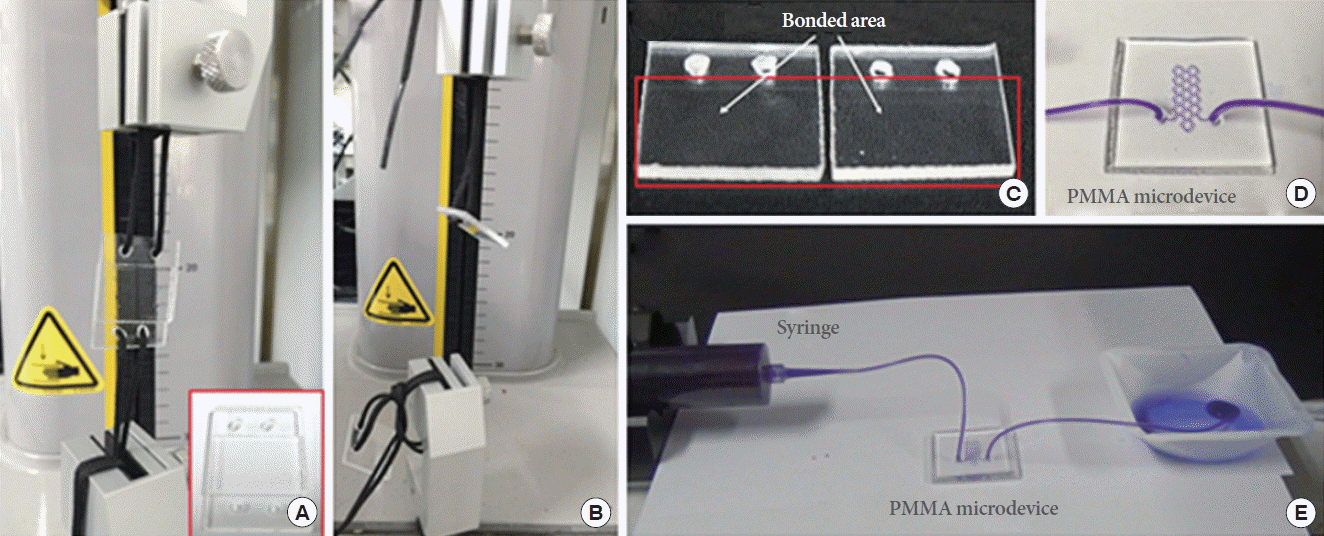
Fig. 5.
(A) Photo of lab-made heater consisting of two aluminum heat blocks and a central controller. (B) Enlarged photo of the two heat blocks shown in panel A. (C) Infrared camera image showing the homogeneous temperature profile on the surfaces of the two heat blocks.
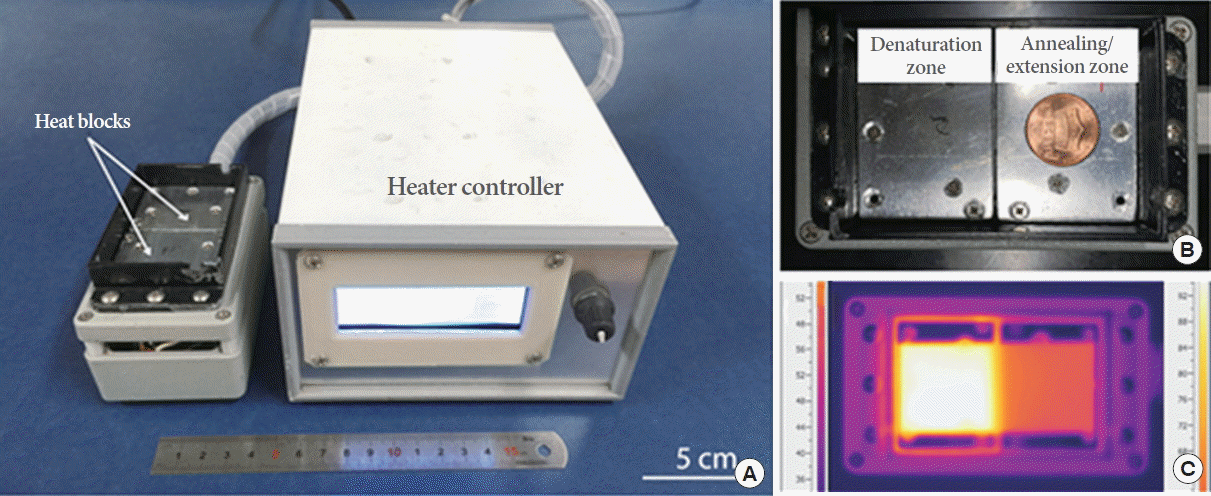
Fig. 6.
Series of images showing the bubble suppression phenomena inside the microchannel under heated conditions for PDMSPDMS Lab-on-a-Chip device, PMMA-PMMA Lab-on-a-Chip device, and PMMA-PMMA Lab-on-a-Chip device coated with epoxy-PDMS on the inner walls of the microchannels. PDMS, poly(dimethylsiloxane); PMMA, poly(methylmethacrylate).
Fig. 7.
(A) Design of the serpentine microchannel for performing continuous-flow PCR. (B) Photo of the PMMA Lab-on-a-Chip device. (C) Photo of the overall system constructed for performing continuous-flow PCR on a PMMA Lab-on-a-Chip device employing a lab-made heater for heating and a syringe pump for sample introduction. (D) Infrared camera image of the Lab-on-a-Chip device showing the temperature profile. (E) Results of 210 bp target amplicons obtained from Escherichia coli O157:H7 using thermal cycler (lane 2) and Lab-on-a-Chip device (lane 3). (F) Results of 689-bp target amplicons obtained from Cronobacter condiment using thermal cycler (lane 2) and the Lab-on-a-Chip device (lane 3). Lane M shows 100-bp DNA size marker, and lane 1 shows the result of the negative control experiment, in both gel images. PCR, polymerase chain reaction; PMMA, poly (methylmethacrylate).
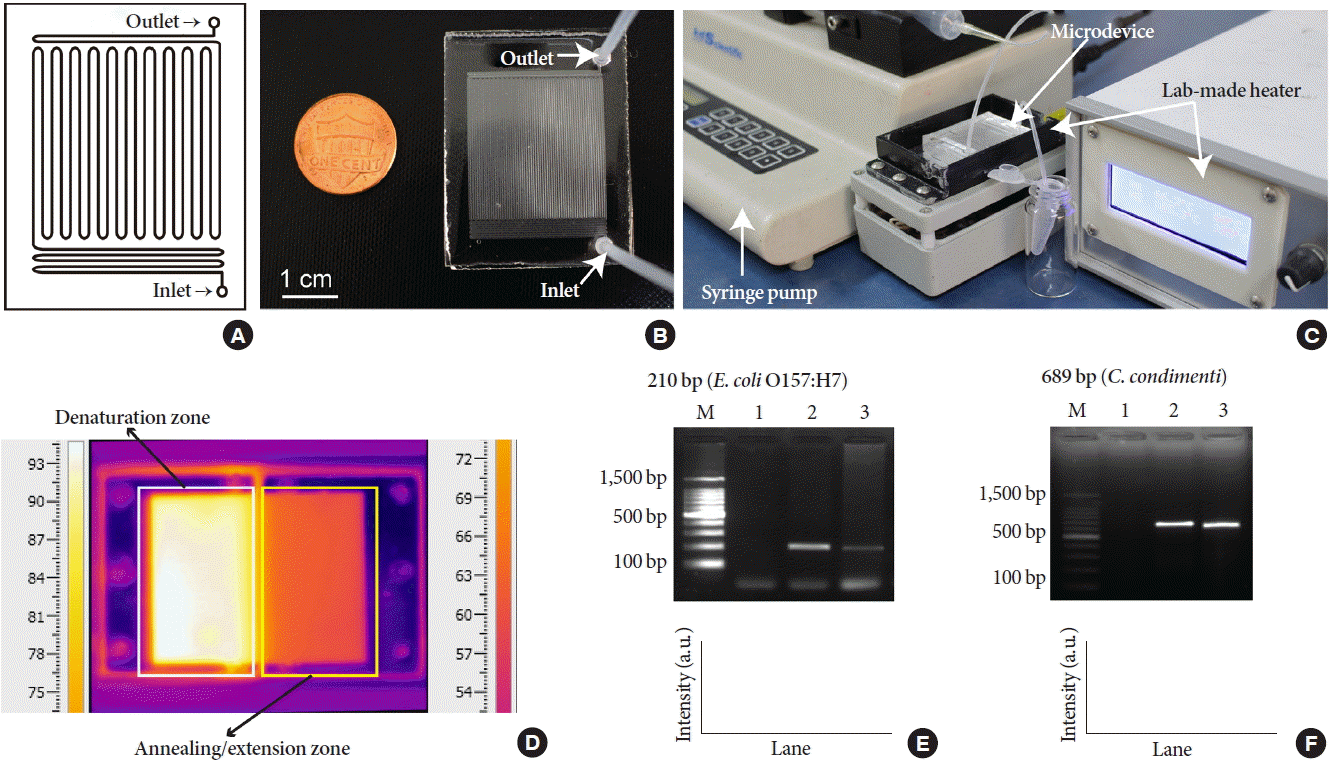
Table 1.
Assignments of the bands after FTIR analyses for pristine PMMA and APTES-coated PMMA