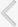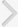|
 |


- Search
| Int Neurourol J > Volume 19(2); 2015 > Article |
|
ABSTRACT
Purpose:
MicroRNAs (miRNAs) in biological fluids are potential biomarkers for the diagnosis and assessment of urological diseases such as benign prostatic hyperplasia (BPH) and prostate cancer (PCa). The aim of the study was to identify and validate urinary cell-free miRNAs that can segregate patients with PCa from those with BPH.
Methods:
In total, 1,052 urine, 150 serum, and 150 prostate tissue samples from patients with PCa or BPH were used in the study. A urine-based miRNA microarray analysis suggested the presence of differentially expressed urinary miRNAs in patients with PCa, and these were further validated in three independent PCa cohorts, using a quantitative reverse transcriptionpolymerase chain reaction analysis.
Results:
The expression levels of hsa-miR-615-3p, hsv1-miR-H18, hsv2-miR-H9-5p, and hsa-miR-4316 were significantly higher in urine samples of patients with PCa than in those of BPH controls. In particular, herpes simplex virus (hsv)-derived hsv1-miR-H18 and hsv2-miR-H9-5p showed better diagnostic performance than did the serum prostate-specific antigen (PSA) test for patients in the PSA gray zone. Furthermore, a combination of urinary hsv2-miR-H9-5p with serum PSA showed high sensitivity and specificity, providing a potential clinical benefit by reducing unnecessary biopsies.
MicroRNAs (miRNAs) are non–protein-encoding RNA regulators and are implicated in numerous biological and developmental processes [1,2]. Approximately 50% of human miRNAs are encoded in genomic regions that are frequently altered in various types of cancers including prostate cancer (PCa) [3-5]. Recently, it has been suggested that miRNAs may be PCa biomarkers since stable miRNAs have been detected in biological fluids such as serum, plasma, and urine [6-8]. In the context of PCa, several putative miRNAs in body fluids have been suggested as diagnostic and prognostic markers [9-11]. However, the complex methodology and low reliability due to relatively small numbers of enrolled patients have limited the clinical significance of these studies.
PCa is the second most commonly diagnosed malignant disease affecting men worldwide, and the incidence and mortality in the Asian population have drastically increased [12,13]. Although total serum prostate-specific antigen (tPSA) levels are the current gold standard for PCa diagnosis in clinical settings, PSA assays often lead to over-diagnosis of patients with an indolent disease. Patients without clinically detectable PCa are often referred to as the “PSA gray zone,” with concentrations of tPSA between 3.0 ng/mL and 10.0 ng/mL. Transrectal ultrasound-guided prostate biopsy is used to facilitate early diagnosis of patients in the PSA gray zone. However, transrectal biopsy is a painful and expensive procedure and has a detection rate of only approximately 30% for PCa patients in the PSA gray zone. Approximately 70% of those patients have no clinical benefits related to this unnecessary and unpleasant procedure. New biomarkers for the early detection of PCa are urgently needed for implementation in clinical practice.
The aim of this study was to examine differences in the expression of urinary cell-free miRNAs between PCa patients and controls with benign prostatic hyperplasia (BPH).
After obtaining approval from the Institutional Review Board of Chungbuk National University Hospital (IRB No. 2006-01-001 and GR2010-12-010). a total of 1,054 urine, 150 serum, and 150 prostate tissue samples from patients with PCa or BPH were used in the study. Written informed consent was obtained from each subject. Patients who underwent radical prostatectomy or palliative transurethral resection of the prostate (TURP) with histologically confirmed primary adenocarcinoma were included. Controls were selected from a database of BPH patients who underwent TURP.
In PCa and BPH patients, first morning voided urine was collected prior to surgery. As for the biopsy patients, spot urine samples were obtained immediately before the procedure. Urine samples were centrifuged at 2,500 rpm for 15 minutes and the supernatant was stored at –80°C until use. Serum samples were obtained on the morning of the operation and stored at –80°C until sample preparation. All prostate tissues were macro-dissected, typically within 15 minutes of surgical resection. All tissue specimens were examined by an experienced senior pathologist (O.J.L.). Gleason grades and TNM 2002 staging were used as prognostic factors. Biopsy cases were selected prospectively in consecutive patients who underwent transrectal ultrasound-guided biopsy.
MicroRNAs were isolated from 500 μL of urine, using the Genolution Urine miRNA Purification Kit (Genolution Pharmaceuticals Inc., Seoul, Korea), and from 200 μL of serum, using the Genolution Serum miRNA Purification Kit (Genolution Pharmaceuticals Inc.) according to the manufacturer’s protocol. The miScript Reverse Transcription Kit (Qiagen Korea, Seoul, Korea) was used to reverse transcribe the miRNAs, using the manufacturer’s recommended protocol.
Total RNA was extracted from urine samples using the RecoverAll Total Nucleic Acid Isolation Kit (Life Technologies, Carlsbad, CA, USA). RNA quantity and integrity were examined with the RNA 6000 Pico Chip Kit (Agilent Technologies, Santa Clara, CA, USA) and Agilent 2100 Bioanalyzer. Profiling of miRNA was performed using the Agilent Human miRNA Microarray Release 16.0 platform, which screens 1,205 human and 144 viral miRNAs.
To quantify miRNA expression derived from the miRNA microarray (GSE54010), real-time polymerase chain reaction (PCR) amplification was performed using the Rotor-Gene Q and miScript PCR Starter Kit (Qiagen Korea). Chemically synthesized RNA oligonucleotides (Cosmo Genetech, Seoul, Korea) corresponding to the target miRNAs were used to generate standard curves. The standard curves ranged from 30 to 3×104 copies. All samples were run in triplicate. The Quant-iT RiboGreen RNA Reagent and Kit (Invitrogen, Grand Island, NY, USA) were used to measure the concentration of total miRNA purified from the samples.
For the miRNA microarray profile, data visualization and analysis were performed using GeneSpring GX ver. 7.3 (Agilent Technologies). Differentially expressed miRNAs were identified using Student t-test with a P-value cutoff of 0.05 and a fold-change threshold of 2.0. One-way analysis of variance (ANOVA) was used to analyze the correlation between miRNA expression and clinicopathological features. Receiver operating characteristic (ROC) curves were used to evaluate the diagnostic performance of the candidate miRNAs, and the optimal cutoff points for candidate markers were determined based on the highest combined sensitivity and specificity for detection in the ROC curve analysis. Spearman correlation coefficient (r) was used to evaluate the correlation between miRNA levels in the matched samples. The statistical analysis was performed using IBM SPSS Statistics ver. 21.0 (IBM Co., Armonk, NY, USA), and a P-value of less than 0.05 was considered statistically significant.
To identify candidate urinary miRNAs specific to PCa, a discovery cohort consisting of fourteen urine samples from PCa patients and five BPH controls was used for miRNA microarray analysis (Table 1). Five miRNAs were selected from the array data, and their expression levels were tested using validation cohort-1. Two herpes simplex virus (hsv)-derived miRNAs were selected for further evaluation using the large validation cohort-2. Simultaneously, expression patterns of candidate miRNAs were compared in matched samples obtained from 99 PCa patients and 51 controls to investigate correlations between tissue, urine, and serum samples. Finally, candidate miRNAs were evaluated as detection markers in 150 urine samples from biopsy patients (validation cohort-3). A workflow and overall study design are shown in Fig. 1.
In order to identify the PCa-specific miRNAs in urine specimens, we used an Agilent miRNA microarray, as described in the Materials and Methods section. Our miRNA profiling analysis suggested that the levels of hsa-miR-615-3p, ebv-miR-BART4, hsv1-miR-H18, hsv2-miR-H9-5p, and hsa-miR-4316 were significantly higher in urine samples from PCa patients than in those from BPH controls (P<0.05). Five miRNAs were selected for further analysis.
Reference markers for miRNAs in human body fluids have not been confirmed to date. Therefore, we attempted to identify accurate reference markers in urine. Candidate urinary reference miRNAs were selected from miRNA microarray data derived from 14 PCa cases (seven localized and seven advanced-stage disease) and five BPH controls. We selected hsa-miR-518a and -3605 because they were detectable in all urine samples from 21 patients and were expressed at similar levels in PCa and BPH patients. In addition, we included hsa-miR-16 and RUN6-2 because they are widely used as reference miRNAs or small RNAs in body fluids or tissues. The four candidate reference miRNAs were validated in 70 urine samples from PCa patients and in 48 samples from BPH controls by real-time PCR. Expression levels were compared between the patient and control groups, and the expression stability was analyzed using the NormFinder program. NormFinder is a Microsoft Excel add-in program that uses an ANOVA-based model to calculate stability values from different subgroups within a panel of candidate genes by combining intra- and intergroup expression variation [14].
Table 2 shows the stability values and expression levels of the candidate markers. The expression of the four candidate miRNAs was significantly different between samples from PCa and BPH patients (P < 0.05). The NormFinder stability values were too high for use as urinary reference markers; therefore, they could not be used in the present study. Instead, miRNA expression was normalized to the total RNA concentration (measured using RiboGreen).
A quantitative reverse transcription-PCR (RT-PCR) analysis was applied to validate the candidate marker miRNAs, and hsa-miR-615-3p, hsv1-miR-H18, hsv2-miR-H9-5p, and hsa-miR-4316 showed significant differences between PCa patients and BPH controls (P<0.05) (Fig. 2). In particular, the levels of hsv1-miR-H18 and hsv2-miR-H9-5p were greatly higher in PCa patients than in BPH controls (P<0.001). An ROC curve analysis was performed to determine whether these urinary miRNAs distinguished between PCa patients and controls. The levels of hsv1-miR-H18, hsv2-miR-H9-5p, hsa-miR-615-3p, and hsa-miR-4316 in the urine showed areas under the curve (AUCs) of 0.750, 0.769, 0.629, and 0.666, respectively. None of these miRNAs showed a difference with respect to clinical stage (T3 or less vs. T4 or any metastasis), Gleason score (≤7 vs. ≥8), or PSA (≤10 ng/mL vs. >10 ng/mL).
The expression levels of five representative miRNAs were measured in matched tissue, serum, and urine samples from 99 PCa patients and 51 BPH controls. We found that hsa-miR-615-3p, hsv1-miR-H18, hsv2-miR-H9-5p, and hsa-miR-4316 showed similar expression patterns in tissue and urine samples. Levels of hsv1-miR-H18 and hsv2-miR-H9-5p were correlated between tissue and serum samples (Fig. 3). Only hsv1-miR-H18 and hsv2-miR-H9-5p expression levels were similarly elevated in tissue, urine, and serum samples. Expression levels of hsv1-miR-H18 and hsv2-miR-H9-5p in tissues showed a better correlation to those in urine than to those in serum samples (Fig. 4).
As an independent approach, we tested two hsv-encoded miRNAs by using a quantitative RT-PCR analysis in a separate cohort (validation cohort-2). Our RT-PCR data provided confirmatory results showing that expression levels of hsv1-miR-H18 and hsv2-miR-H9-5p were significantly different between PCa and BPH control samples (P<0.001). An ROC analysis, a conventional tool for diagnostic test evaluations, was performed to determine how well hsv1-miR-H18 and hsv2-miR-H9-5p expression levels segregate PCa patients from controls. In the ROC analysis, the levels of hsv1-miR-H18 in the urine showed an AUC of 0.772, with a sensitivity of 66.5% and a specificity of 74.1% (Fig. 5A, left). The levels of hsv2-miR-H9-5p showed an AUC of 0.777, with a sensitivity of 70.2% and a specificity of 72.0% (Fig. 5A, middle). For patients within the PSA gray zone (tPSA, 3–10 ng/mL), the AUCs for hsv1-miR-H18 and hsv2-miR-H9-5p (0.761 and 0.738, respectively) were much higher than that of tPSA (0.613) (Fig. 5B). These findings suggest that hsv1-miR-H18 and hsv2-miR-H9-5p provide better diagnostic performance than the serum tPSA test, a gold standard diagnostic method for PCa, for patients in the PSA gray zone (Fig. 5B, right).
Diagnostic performances of hsv1-miR-H18 and hsv2-miR-H9-5p were re-evaluated for the patients who underwent transrectal prostate biopsy. The AUCs for hsv1-miR-H18 and hsv2-miR-H9-5p were 0.639 and 0.760, respectively. These data suggest that a quantitative RT-PCR analysis of urinary hsv2-miR-H9-5p can provide strong discrimination ability, comparable to that of serum tPSA levels (AUC=0.710) (Fig. 5C). We then hypothesized that PCa patients could be efficiently diagnosed using the combined signature of hsv2-miR-H9-5p and tPSA. The cutoff values of 109.87×102 copies/miRNA for hsv2-miR-H9-5p and 3.0 ng/mL for tPSA were used for this analysis. Based on this analysis, 53.1% subjects (34 of 64) with high hsv2-miR-H9-5p had PCa, while 86.0% of subjects (74 of 86) with low hsv2-miR-H9-5p did not have PCa, suggesting that high expression of hsv2-miR-H9-5p is strongly associated with PCa. These findings suggest that a test measuring the combined signature of urinary hsv2-miR-H9-5p and serum tPSA may be a substitute method that can minimize the use of unnecessary transrectal prostate biopsy.
The current study provided multiple layers of evidence suggesting that two “cell-free” virus-encoded miRNAs (hsv1-miR-H18 and hsv2-miR-H9-5p) are detected in urine and can serve as strong diagnostic biomarkers for PCa. In particular, hsv2-miR-H9-5p had a comparable detection power to that of serum tPSA in patients who underwent transrectal biopsy, and the combination of hsv2-miR-H9-5p and serum PSA could provide clinical data to reduce unnecessary biopsy.
The association of HSV type 2 infection with an increased PCa risk was recently revealed by a meta-analysis, although the mechanism underlying viral carcinogenesis remains elusive. Not only HSV, but also Epstein-Bar virus and Marek disease viruses are associated with tumorigenesis and encode miRNAs. However, tumorigenesis is not the primary “aim” of these viruses, and it is an unfortunate consequence of their ability to alter the cell cycle, prevent cell death, and avoid immune responses in host cells [15]. Since Pfeffer et al. [16] first identified five EBV-encoded pre-miRNAs in B95-8 cells in 2004, more than 250 virus-encoded miRNAs have been identified. Virus-encoded miRNAs are grouped into two classes: analogues of host miRNAs and virus-specific miRNAs [17]. Virus-encoded miRNAs mimic specific host miRNAs and negatively regulate transcription via the same target docking sites as their counterpart host miRNAs [18,19], and regulate host cells in several ways: (1) by regulation of the latent-lytic switch; (2) by supporting viral replication through promoting cell survival, proliferation, and/or differentiation; and (3) by modulating immune responses [17]. Given that the evidence supporting an association between PCa and inflammation caused by bacteria and viruses [20], it is possible that hsv1-miR-H18 and hsv2-miR-H9-5p play a role in tumorigenesis. Furthermore, given that the increased levels of hsv1-miR-H18 and hsv2-miR-H9-5p identified early-stage PCa patients in the PSA gray zone, they may provide clinical benefits for early detection of PCa.
A number of studies report promising results regarding the use of putative miRNAs in blood or urine as diagnostic and prognostic markers for PCa. Our present study took novel approaches and to the results have significant clinical implications. Our study was performed on 593 cases and 459 controls, and on six independent cohorts in total, which was the largest study to investigate putative miRNAs in body fluids from PCa patients. Additionally, we introduced an established protocol to extract RNA (including miRNAs and small RNAs) that requires only 0.5 mL of voided urine, which is of clinical significance. We measured the levels of cell-free miRNA in urine supernatants rather than in urine sediments, which contain mixed cell populations including immune, blood, kidney, and bladder cells. Two studies have suggested that urinary miRNAs can be used as biomarkers for PCa [10,21]. Bryant et al. [10] suggested that miR-107 and miR-574-3p are present at higher levels in the urine (collected after transrectal digital massage) of PCa patients than in that of healthy controls. Srivastava et al. [21] reported that miR-205 and -214 are downregulated in the urine of PCa patients and discriminate patients from healthy controls. They selected miRNAs from a tissue miRNA array and examined their levels in urine samples using RNU48 for normalization, which is a different approach from that used in our study. Novel findings from our study included the better diagnostic performance of hsv1-miR-H18 and hsv2-miR-H9-5p than tPSA levels in patients within the PSA gray zone. Although numerous diagnostic markers have been developed and tested in an attempt to increase diagnostic power, these markers show poor reproducibility and the methods are too complex to use in the clinical setting.
Collectively, our findings in this study suggest that virus-encoded miRNAs may be applied as a diagnostic marker for PCa in clinical practice. To the best of our knowledge, this is the first study reporting an association between virus-encoded miRNAs and PCa.
ACKNOWLEDGEMENTS
The biospecimens for this study were provided by the Chungbuk National University Hospital, a member of the National Biobank of Korea, which is supported by the Ministry of Health, Welfare, and Family Affairs. All samples derived from the National Biobank of Korea were obtained with informed consent under Institutional Review Board-approved protocols.
The authors wish to thank Ms. Eun-Ju Shim from the National Biobank of Korea at Chungbuk National University Hospital for the sample preparations and her excellent technical assistance.
NOTES
Grant Support
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (No. NRF-2014R1A2A1A09006983) and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2012R1A1A4A01008753 and
2013R1A1A2004740).
REFERENCES
2. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. PMID: 14744438


3. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004;101:2999-3004. PMID: 14973191



4. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. PMID: 15944708


5. Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 2005;353:1768-71. PMID: 16251533


6. Kim SM, Kang HW, Kim WT, Kim YJ, Yun SJ, Lee SC, et al. Cellfree microRNA-214 from urine as a biomarker for non-muscle-invasive bladder cancer. Korean J Urol 2013;54:791-6. PMID: 24255763



7. Westermann AM, Schmidt D, Holdenrieder S, Moritz R, Semjonow A, Schmidt M, et al. Serum microRNAs as biomarkers in patients undergoing prostate biopsy: results from a prospective multi-center study. Anticancer Res 2014;34:665-9. PMID: 24510997

8. Yun SJ, Jeong P, Kim WT, Kim TH, Lee YS, Song PH, et al. Cellfree microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol 2012;41:1871-8. PMID: 22961325


9. Shen J, Hruby GW, McKiernan JM, Gurvich I, Lipsky MJ, Benson MC, et al. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 2012;72:1469-77. PMID: 22298119



10. Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer 2012;106:768-74. PMID: 22240788



11. Delgado PO, Alves BC, Gehrke Fde S, Kuniyoshi RK, Wroclavski ML, Del Giglio A, et al. Characterization of cell-free circulating DNA in plasma in patients with prostate cancer. Tumour Biol 2013;34:983-6. PMID: 23269609


12. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300. PMID: 20610543


13. Jung KW, Won YJ, Park S, Kong HJ, Sung J, Shin HR, et al. Cancer statistics in Korea: incidence, mortality and survival in 2005. J Korean Med Sci 2009;24:995-1003. PMID: 19949651



14. Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004;64:5245-50. PMID: 15289330


15. Zhao Y, Xu H, Yao Y, Smith LP, Kgosana L, Green J, et al. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathog 2011;7:e1001305. 
16. Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science 2004;304:734-6. PMID: 15118162


17. Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol 2010;64:123-41. PMID: 20477536



19. Kincaid RP, Burke JM, Sullivan CS. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci USA 2012;109:3077-82. PMID: 22308400



20. De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer 2007;7:256-69. PMID: 17384581



21. Srivastava A, Goldberger H, Dimtchev A, Ramalinga M, Chijioke J, Marian C, et al. MicroRNA profiling in prostate cancer--the diagnostic potential of urinary miR-205 and miR-214. PLoS One 2013;8:e76994. PMID: 24167554



Fig. 1.
Overview of the study design. A heatmap representing microRNA (miRNA) microarray data suggested that the miRNA signature segregates prostate cancer (PCa) from benign prostatic hyperplasia (BPH) controls (top right).
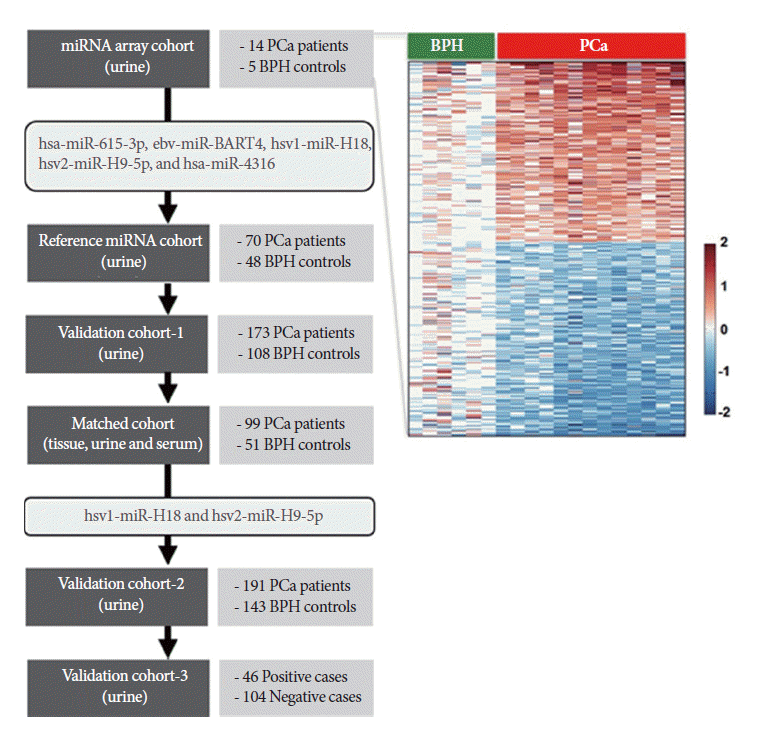
Fig. 2.
Box plots showing the expression of five urinary microRNAs (miRNAs) in benign prostatic hyperplasia (BPH) controls (n=108) and prostate cancer (PCa) patients (n=173). Of these, hsa-miR-615-3p (A), hsv1-miR-H18 (C), hsv2-miR-H9-5p (D), and hsa-miR-4316 (E) showed significant differences between BPH controls and PCa patients. However, the expression of ebvmiR- BART4 (B) was different from that observed in the array data.
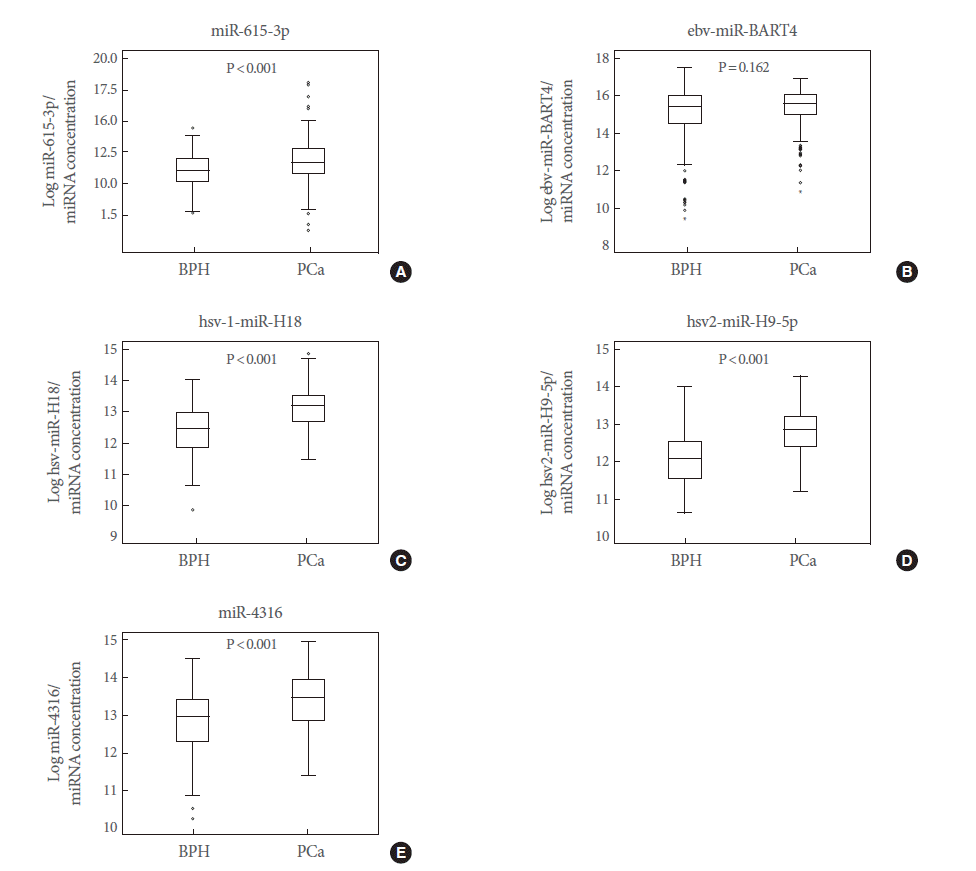
Fig. 3.
Expression patterns of five candidate miRNAs in matched prostate tissue, urine, and serum samples. Of these, hsa-miR-615-3p (A), hsv1-miR-H18 (C), hsv2-miR-H9-5p (D), and hsa-miR-4316 (E) showed similar expression patterns in tissue and urine samples, while hsv1-miR-H18 (C) and hsv2-miR-H9-5p (D) were correlated between tissue and serum samples. More importantly, only hsv1-miR-H18 and hsv2-miR-H9-5p expression levels were similarly elevated in tissue, urine, and serum samples. miRNA, microRNA; BPH, benign prostatic hyperplasia; PCa, prostate cancer. Expression patterns of five candidate miRNAs in matched prostate tissue, urine, and serum samples. Of these, hsa-miR-615-3p (A), hsv1-miR-H18 (C), hsv2-miR-H9-5p (D), and hsa-miR-4316 (E) showed similar expression patterns in tissue and urine samples, while hsv1-miR-H18 (C) and hsv2-miR-H9-5p (D) were correlated between tissue and serum samples. More importantly, only hsv1-miR-H18 and hsv2-miR-H9-5p expression levels were similarly elevated in tissue, urine, and serum samples. miRNA, microRNA; BPH, benign prostatic hyperplasia; PCa, prostate cancer.
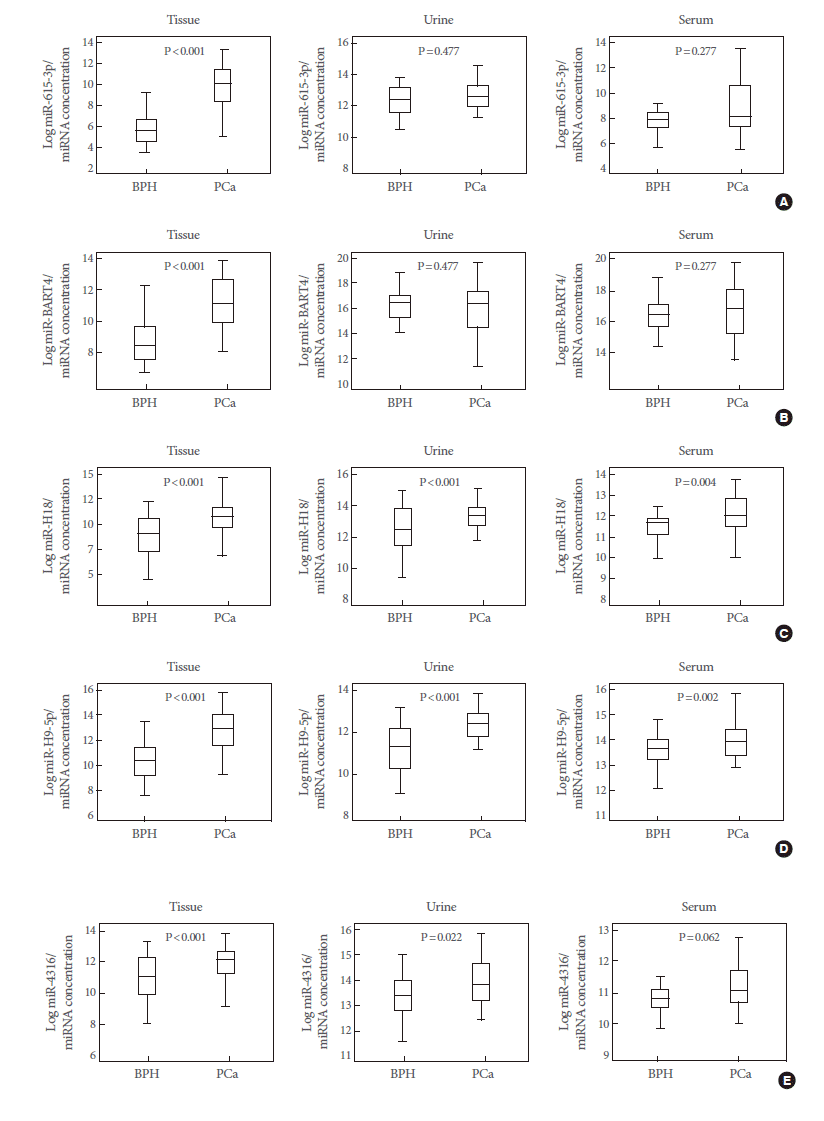
Fig. 4.
(A, B) Correlation between hsv1-miR-H18 and hsv2-miR-H9-5p levels in matched tissue, urine, and serum samples. Black circles indicate prostate cancer patients and open circles indicate benign prostatic hyperplasia controls. There was a significant correlation between hsv1-miR-H18 (A) and hsv2-miR-H9-5p (B) levels in tissue and urine samples (Spearman correlation=0.369, P<0.001, and Spearman correlation=0.552, P<0.001, respectively). On the other hand, there was only a weak correlation between the levels in tissue and serum samples. miRNA, microRNA.
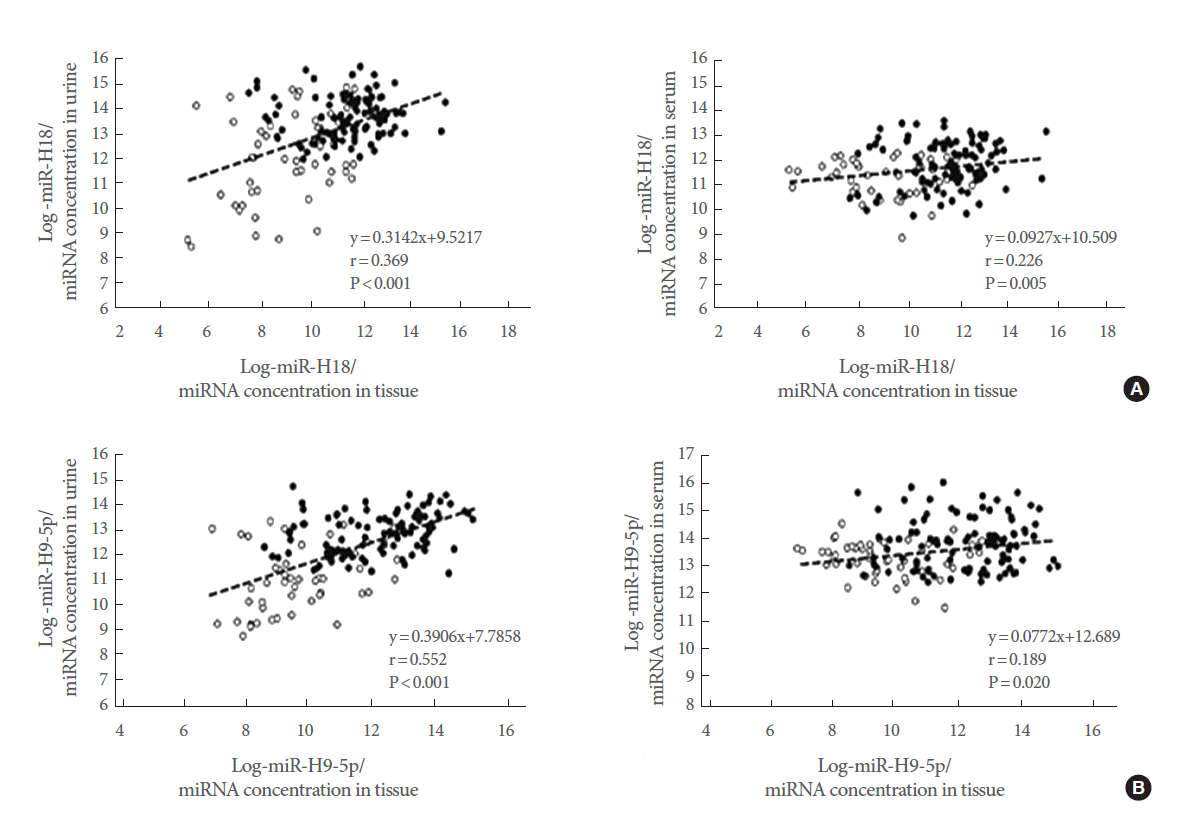
Fig. 5.
Diagnostic performance of urinary hsv1-miR-H18 and hsv2-miR-H9-5p compared with serum prostate-specific antigen (PSA) levels. (A) Receiver operating characteristic (ROC) curves for all patients (n=191) and controls (n=143). (B) ROC curves for patients within the PSA gray zone (3–10 ng/mL). (C) ROC curves for biopsy cases. The optimal cutoff points were determined based on the ROC curve analysis that yielded the highest combined sensitivity and specificity for detection.

Table 1.
Clinical characteristics of the patients and controls in the study
Table 2.
NormFinder stability values and expression levels of candidate urinary reference markers in PCa patients and BPH controls
| miRNA ID | Stability valuea) | BPH controls (n=48) | PCa cases (n=70) | P-value |
|---|---|---|---|---|
| hsa-miR-518a | 408,991.50 | 4,098.6 (3,566.3–10,639.7) | 2,670.6 (1,904.3–4,097.3) | < 0.001 |
| hsa-miR-3605 | 647,977.50 | 8,842.4 (2,952.6–44,122.7) | 2,781.7 (1,015.4–21,381.8) | 0.033 |
| hsa-miR-16 | 119,927.70 | 1,030.7 (525.7–2,048.6) | 619.2 (320.0–1,018.6) | 0.040 |
| RUN6-2 | 61,037.20 | 752.8 (302.1–2,537.1) | 230.5 (92.6–585.5) | < 0.001 |