Association Study of Polymorphisms of Epidermal Growth Factor and Epidermal Growth Factor Receptor With Benign Prostatic Hyperplasia in a Korean Population
Article information
Abstract
Purpose
Recent studies have suggested that specific single-nucleotide polymorphisms (SNPs) contribute to the clinical features of benign prostatic hyperplasia (BPH). In this study, we investigated the relationships of genetic polymorphisms of the epidermal growth factor (EGF) gene and the epidermal growth factor receptor (EGFR) gene with BPH.
Methods
A total of 218 patients with BPH were enrolled in this study. We evaluated the relationship between eight SNPs in the EGF and EGFR genes and prostate volume, prostate-specific antigen (PSA), and International Prostate Symptom Score of BPH patients. Each SNP was genotyped by direct sequencing. Statistical analysis applying codominant, dominant, recessive, and log-additive models was performed via logistic regression.
Results
The rs11568943 and rs11569017 SNPs in the EGF gene showed significant associations with prostate volume (rs11568943: P=0.038 in the log-additive model, P=0.024 in the allele distribution; rs11569017, P=0.031 in the dominant model, P=0.028 in the log-additive model, P=0.020 in the allele distribution). Additionally, the rs3756261, rs11568943, and rs11569017 SNPs of the EGF gene and the rs2293347 SNP of the EGFR gene were associated with PSA levels (P<0.05 in each model, respectively).
Conclusions
These results suggest that the EGF gene may affect prostate volume. In addition, the EGF and EGFR genes may be associated with PSA levels in patients with BPH.
INTRODUCTION
Benign prostatic hyperplasia (BPH) is a pathologic process that is one of the causes of lower urinary tract symptoms (LUTS) in aging men. BPH may cause symptoms including LUTS, benign prostatic enlargement, and bladder outlet obstruction. BPH has similar risk factors to those of prostate cancer [1]. Epithelial cell proliferation in prostate tissue is a major pathologic feature of BPH [2]. Epidermal growth factor (EGF) is linked to the growth and differentiation of epithelial cells. The relationship between EGF and prostate cancer has been previously studied. EGF, vascular endothelial growth factor, fibroblast growth factor 2, transforming growth factor beta 1, and insulin-like growth factor 1 are known to influence the development of both prostate cancer and BPH [3,4], and both conditions share risk factors. However, the relationship between BPH and EGF remains unclear [1].
Evidence has emerged suggesting that the action of EGF is involved in prostate cell growth. EGF expression is promoted by exposure to androgens, such as testosterone and dihydrotestosterone [5-7]. EGF may cause elevations in E-cadherin, which may promote the phosphorylation of epidermal growth factor receptor (EGFR) and cell proliferation signals [8]. Recent studies have shown that the inhibition of EGFR was linked to decreased proliferation signals to prostatic epithelial cell lines [9].
Although several studies have investigated EGF, only 1 polymorphism in the EGF gene has been reported to affect the development of prostate cancer. Teixeira et al. [10,11] reported that the EGF +61G>A polymorphism may contribute to prostate cancer susceptibility and androgen insensitivity. Studies of the EGFR gene have shown the features or development of prostate cancer to be associated with various exon mutations [12,13] and single-nucleotide polymorphisms (SNPs) including rs17172432 [14], rs6964705 in the EGFR gene combined with rs1401862 in the matrix metallopeptidase 16 (MMP16) gene [15], and rs884419 [16].
Although the above studies support a relationship between EGF signals and proliferative prostate disease, no study has investigated the relationship of BPH with the EGF and EGFR genes. In the present study, we investigated the relationship between SNPs of the EGF and EGFR genes and the clinical features of BPH in a Korean population.
MATERIALS AND METHODS
Study Subjects
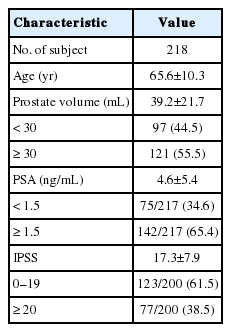
All subjects were recruited from the Kyung Hee University Medical Center of Kyung Hee University in Seoul, Korea. This study was approved by Institutional Review Board of Kyung Hee University Medical Center in 2009 (KMC IRB 0913-03). A total of 218 BPH patients diagnosed by a physician were selected (Table 1). The criteria for diagnosing BPH included prostate weight (>20 g) and LUTS. The prostate-specific antigen (PSA) level in the serum of BPH patients was tested and prostate volume was measured using transrectal ultrasonography by urologists. LUTS were quantified using the International Prostate Symptom Score (IPSS). The subjects were dichotomized according to the criteria used in several multicenter studies: low (0–19) and high (≥ 20) IPSS score, low (<1.5 ng/mL) and high (≥1.5 ng/mL) PSA level, and small (<30 mL) and large (≥30 mL) prostate volume. Voiding symptoms were classified using the IPSS score as mild (0–7), moderate (8–19), or severe (20–35).
The exclusion criteria were prostate cancer, neurogenic bladder, urinary tract infection, uncontrolled diabetes mellitus, and cardiovascular disease.
Written informed consent was obtained from all participants. Blood samples were collected in tubes containing ethylenediaminetetraacetic acid as an anticlotting factor and stored at −20°C until use. Genomic DNA was extracted using a blood extraction kit (Roche, Indianapolis, IN, USA).
SNP Selection and Genotyping
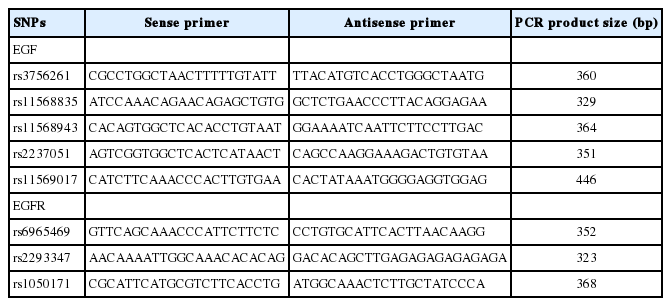
The National Center for Biotechnology Information SNP database was searched to select SNPs of the EGF and EGFR genes for study (http://www.ncbi.nlm.nih.gov/SNP, BUILD 141). The criteria for the selection of exonic SNPs and promoter SNPs in each gene were the following: (1) >10% minor allele frequency, (2) >0.1 heterozygosity, (3) known genotype frequencies in the Asian population, and (4) previous studies. We ultimately selected 5 SNPs of the EGF gene (rs3756261, -1744 A/G; rs11568835, -1380 G/A; rs11568943, Arg431Lys; rs2237051, Met708Ile; and rs11569017, Asp784Val) and 3 SNPs of the EGFR gene (rs6965469, -2004 C/T; rs2293347, Asp994Asp; and rs1050171, Gln787Gln). Additionally, the genotype of each SNP was determined through direct sequencing after polymerase chain reaction (PCR). PCR primers are shown in Table 2. Sequence data were analyzed using SeqManII software (v2.3; DNAATAR Inc., Madison, WI, USA).
Statistical Analysis
The derivation of tested SNPs in Hardy-Weinberg equilibrium was evaluated using SNPstats (http://bioinfo.iconcologia.net/snpstats/start.htm). Differences in the genotypes and alleles of each SNP were analyzed by SNPstat and IBM SPSS Statistics ver. 20.0 (IBM Co., Armonk, NY, USA). The chi-square test and logistic regression with codominant, dominant, recessive, and log-additive models were utilized to analyze the association between tested polymorphisms and prostate volume, PSA level, or IPSS [17,18]. The linkage disequilibrium (LD) block and haplotypes between pairs of SNPs in each gene were tested using Haploview ver. 4.2 (Daly Laboratory, Cambridge, MA, USA). P-values <0.05 were considered to indicate statistical significance.
RESULTS
The demographic and biochemical characteristics of the participants are shown in Table 1. We analyzed the relationships between polymorphisms of the EGF and EGFR genes and BPH. The BPH patients were dichotomized according to prostate volume, IPSS score, and PSA level [19,20]. The genotype and allele distributions of the tested SNPs are shown in Tables 3 and 4 for each group. The derivation of the tested SNPs remained in Hardy-Weinberg equilibrium (rs3756261, P =0.35; rs11568835, P=0.30; rs11568943, P=0.23; rs2237051, P=0.51; rs11569017, P=0.28; and rs2293347, P=0.48 in the EGF gene; rs1050171, P=0.88; and rs6965469, P=0.65 in the EGFR gene).
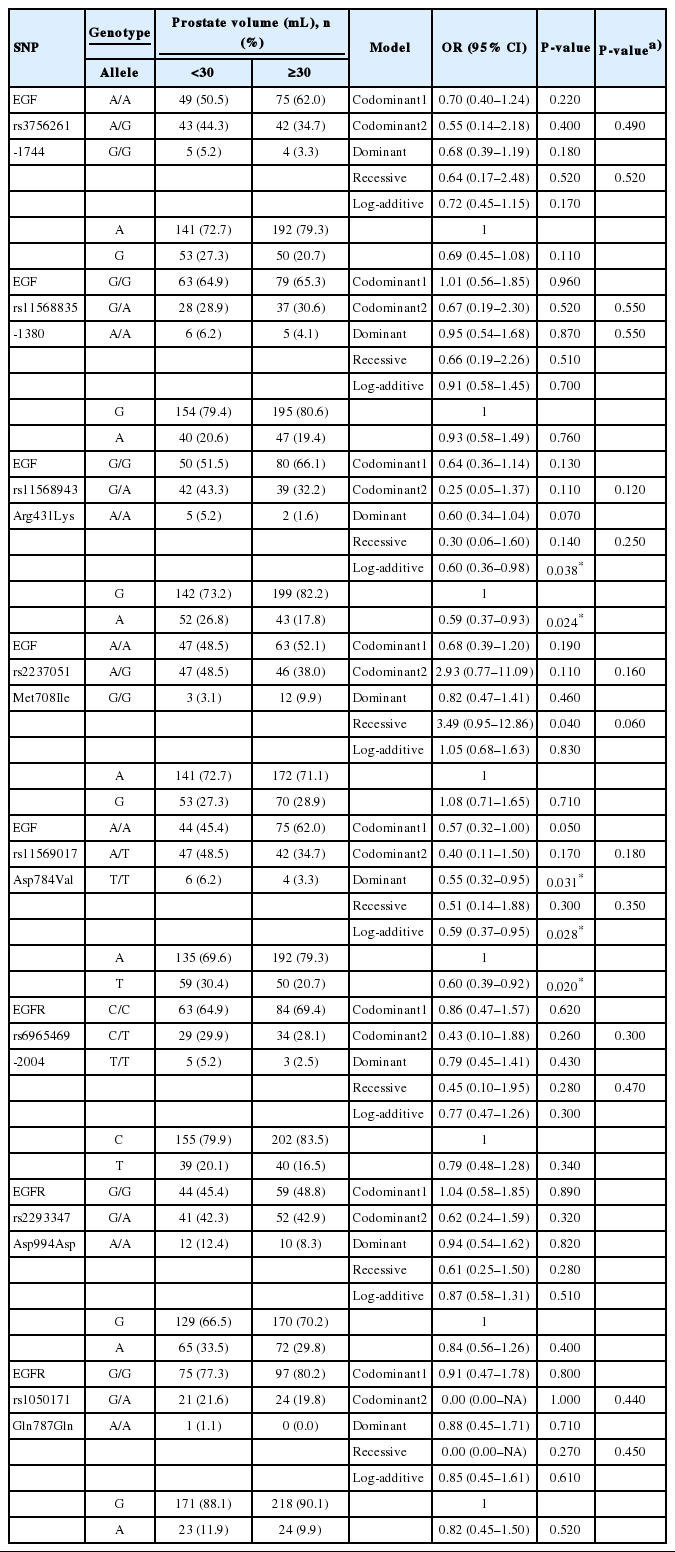
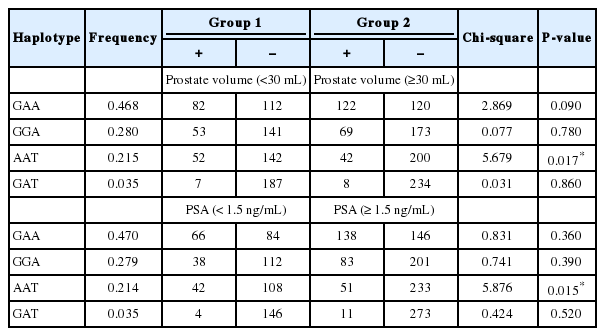
First, we analyzed the relationship between polymorphisms of the EGF and EGFR genes and prostate volume. Codominant, dominant, recessive, and log-additive models were applied for statistical analysis. We found that EGF polymorphisms were associated with prostate volume in BPH patients (Table 3). The distributions of the G allele of rs11568943 and the A allele of rs11569017 in the EGF gene were significantly higher in patients with a prostate volume ≥30 mL than in patients with a prostate volume <30 mL (rs11568943, P=0.024 and rs11569017, P=0.02). The genotype distributions of EGF polymorphisms also showed significant associations with prostate volume (rs11568943, P =0.038 in the log-additive model [G/G vs. A/G vs. A/A]; rs11569017, P=0.031 in the dominant model [A/A vs. A/T+T/T] and P=0.028 in the log-additive model [A/A vs. A/T vs. T/T]). In order to analyze haplotypes, we tested the LD block between paired SNPs in each gene. One LD block was found in the EGF gene among 3 SNPs (rs11568943, rs2237051, and rs11569017), and the LD block was strong (rs11568943 and rs2237051, D’=1.0, r2=0.109; rs11568943 and rs11569017, D’ =0.985, r2=0.812; rs2237051 and rs11569017, D’=1.0, r2=0.131). Four haplotypes were found in the LD block of the EGF gene. Among these haplotypes, a significant difference were found according to AAT haplotype frequency in prostate volume (<30 mL or ≥30 mL) (P=0.017) (Table 5).
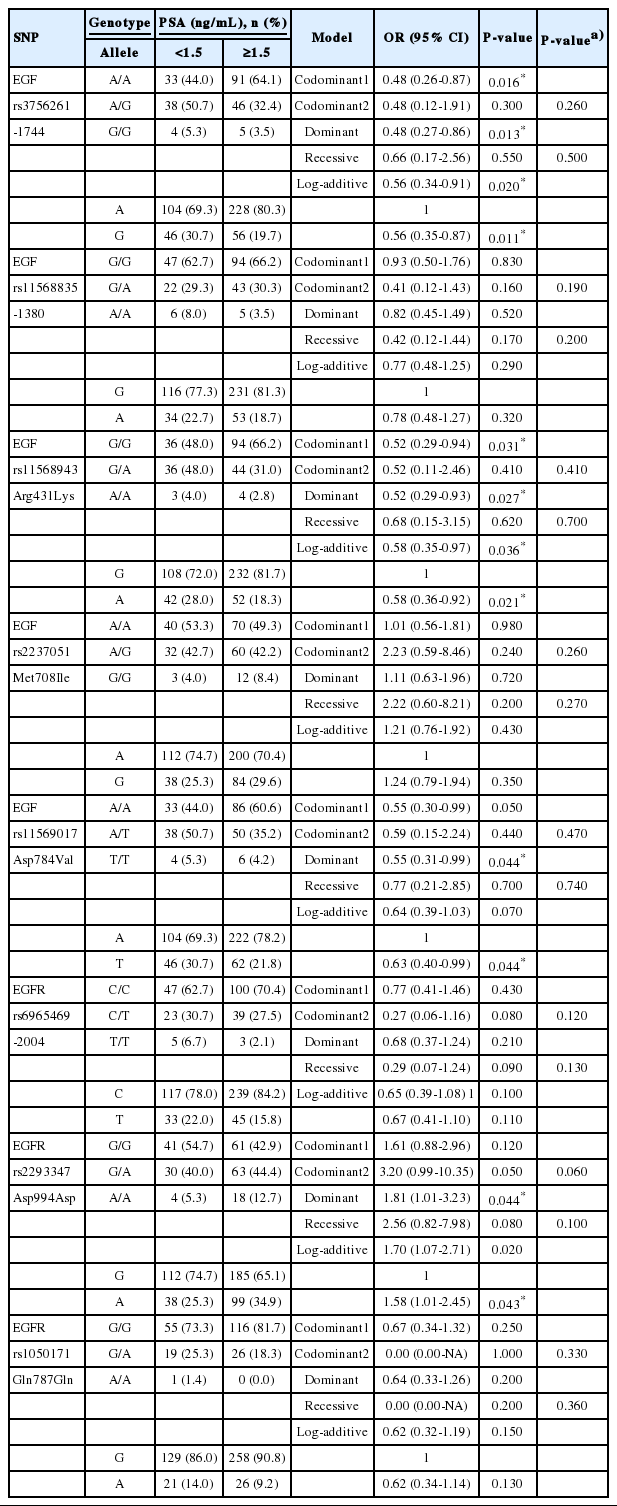
Second, we evaluated the relationship between polymorphisms of the EGF and EGFR genes and PSA levels, and found significant associations (Table 4). Three SNPs (rs3756261, -1744 A/G; rs11568943, Arg431Lys; and rs11569017, Asp784Val) in the EGF gene showed significant associations. The distributions of the major alleles of rs3756261 and rs11568943 were higher in patients with a PSA level ≥1.5 ng/mL than in patients with PSA levels <1.5 ng/mL (rs3756261, P =0.011; rs11568943, P=0.021). The genotype distributions of EGF polymorphisms also displayed significant differences (rs3756261, P=0.016 in the codominant 1 model [A/A vs. A/G], P=0.013 in the dominant model [A/A vs. A/G+G/G], P=0.020 in the log-additive model [A/A vs. A/G vs. G/G]; rs11568943, P=0.031 in the codominant 1 model [G/G vs. G/A], P=0.027 in the dominant model [G/G vs. G/A+A/A], P=0.036 in the log-additive model [G/G vs. G/A vs. A/A]; rs11569017, P=0.044 in the dominant model [A/A vs. A/T+T/T]). Additionally, rs2293347 in the EGFR gene showed a relationship with PSA (P=0.044 in the dominant model [G/G vs. G/A+A/A], P=0.020 in the log-additive model [G/G vs. G/A vs. A/A], and P=0.043 in the allele distribution]. In the haplotype analysis, a significant association was found between the AAT haplotype in the EGF gene and PSA levels (P=0.015) (Table 5).
However, we did not find any significant associations between polymorphisms of the EGF and EGFR genes and IPSS.
DISCUSSION
The pathogenesis of BPH is still unknown. However, several studies have reported that specific polymorphisms in various genes contribute to the pathogenesis of BPH [21]. The most representative genes of this type is the androgen receptor (AR) gene. AR is a transactivation factor that depends on the binding of steroid hormones. It has an important role in the proliferation and differentiation of prostate cells [22]. Polymorphic variations are present in the AR gene. It has been suggested that the presence of a higher number of polymorphic GGC repeats in the AR gene is associated with an increased risk of developing BPH [23]. Short CAG alleles may be a genetic factor that promotes the growth of BPH [24].
The relationship between EGF and EGFR polymorphisms and the clinical features of BPH has not been previously investigated. In the current study, associations between SNPs in the EGF and EGFR genes and BPH were evaluated. The prostate volume of BPH patients was associated with the EGF SNPs rs11568943 and rs11569017, and PSA levels in BPH patients were associated with the EGF SNPs rs11568943, rs11569017, and rs3756261 and the EGFR SNP rs2293347.
In previous studies of the EGF and EGFR genes, the rs 11568943 SNP was significantly associated with preeclampsia [25], psoriatic arthritis [26], and gastric cancer [27]. The rs11569017 SNP was associated with the risk of hepatitis B virus-related hepatocellular carcinoma [28]. The rs3756261 SNP was also showed a significant association with a higher risk of developing preeclampsia [25]. Among the EGFR SNPs, the rs2293347 SNP is associated with chemotherapeutic response, lung cancer treated with gefitinib, and airway hyperresponsiveness.
EGF binding induces the dimerization of EGFR and the activation of downstream signaling pathways involved in regulating cellular proliferation, differentiation, and survival [29]. This EGF-EGFR ligand-receptor complex stimulates cell proliferation [30]. EGF protects epithelial cells against Fas-induced apoptosis [31], and EGFR plays an essential role in the morphogenesis of mammary glands [32]. EGF controls myoepithelial cell differentiation in mammary gland cultures [33] and EGFR is closely linked to smooth cell apoptosis [34]. The prostate gland consists of the peripheral zone, central zone, and transition zone. The transition zone, which contains smooth muscle cells and myoepithelial cells, is responsible for BPH [35]. As described above, EGF and EGFR may be related to various aspects of epithelial and muscular proliferation, which could affect BPH.
In summary, no associations between the EGF and EGFR genes and the clinical features of BPH have previously been reported. We found for the first time that 2 SNPs (rs11568943 and rs11569017) and the AAT haplotype in the EGF gene may affect prostate volume, and that 3 SNPs (rs11568943, rs11569017, and rs3756261) and the AAT haplotype in EGF gene, as well as 1 SNP in the EGFR gene (rs2293347), may affect PSA levels in BPH patients. However, this study has limitations such as sample size, ethnic differences, lack of control subjects, and interaction with environmental factors. To confirm our results, a casecontrol study in another population with a larger sample size is needed, as well as a meta-analysis.
Notes
Research Ethics
This study was approved by Institutional Review Board of Kyung Hee University Medical Center in 2009 (KMC IRB 0913-03).
Conflict of Interest
No potential conflict of interest relevant to this article was reported.