The Urinary Tract Microbiome in Male Genitourinary Diseases: Focusing on Benign Prostate Hyperplasia and Lower Urinary Tract Symptoms
Article information
Abstract
The human body is sterile during gestation; however, but during and after birth, the entire body surface becomes host to an enormous variety of microorganisms. Urine in the urinary tract was once considered sterile based on the lack of cultured microorganisms. Many recent studies have revealed evidence of microorganisms in human urine in the absence of clinical infection. Sequencing methods and analytical techniques are rapidly evolving to improve the ability to detect bacterial DNA and living bacteria and to understand the microbiota of the urinary tract. In women, fascinating evidence associates urinary tract microbiota with lower urinary tract symptoms. However, in men, the relevance of urinary tract microbiota in low urinary tract symptoms and prostate disease has not been established. In this review, we highlight a recent study that increases our ability to understand the urinary tract microbiota in men with lower urinary tract symptoms.
INTRODUCTION
The human body is sterile until birth; however, during birth and the first few years of life, our bodies are colonized by various microorganisms that form a community known as human microbes [1-3]. Microorganisms can include bacteria, archaea, fungi, and protozoa [4,5]. Our microbial community contains 10 times more cells than the rest of the human body, and it is estimated that the human body has more than 1012 microbes with much more genetic material than human gene [1,4]. The ecological community of symbiotic and pathogenic microorganisms living in our body is the term “microbiota,” the genetic material of these microorganisms is the term “microbiome.” [5] However, the microbial community of the human body has changed and the relationship is not perfect [5]. Changes in the microbial population can result from repetitive or coexisting stressors, including age, diet, exercise, smoking, drugs, illness, and other environmental factors [6-10]. These changes in the microflora, often called dysbiosis, are associated with diseases such as obesity, diabetes, multiple sclerosis, allergies, asthma, heart disease, cancer, rheumatoid arthritis, and inflammatory bowel disease [5,11]. Advances in molecular biology techniques and cultivation methods have enabled the detection of certain microbial communities associated with some body parts previously thought to be sterile, including the urinary tract [12-16]. To date, most studies have focused entirely on the microbiota of the genitourinary tract. The discovery of the bacterial community and the investigation of their role in urological diseases led to the proposition of new factors that could influence the pathophysiology and management of these common urological conditions [17,18]. The pathophysiological mechanisms of prostate disease are incompletely characterized [19-21]. Chronic inflammation of the prostate has been implicated as a possible common cause in benign prostatic hyperplasia (BPH), and chronic prostatitis (CP)/chronic pelvic pain syndrome (CPPS), although the cause of inflammation remains controversial [19-21]. According to recent studies, the microbiota can affect prostate inflammation in connection with benign prostate diseases such as CP/CPPS and BPH as well as prostate cancer [20,21]. In this review, we summarize the main studies on urinary microflora in urinary tract diseases in men.
DISCOVERING URINARY MICROBIOTA IN HEALTHY MEN
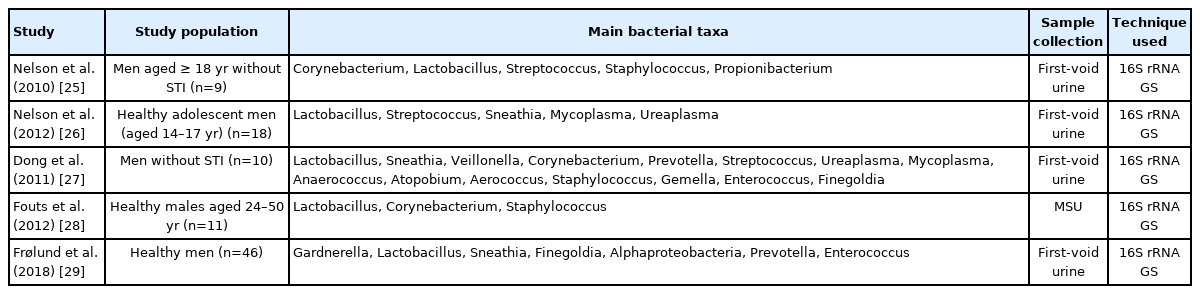
The Human Microbial Community Project (HMP) was established in 2008 to characterize the human microbial community and analyze its role in human health and disease [22]. Five body parts (gastrointestinal, mouth, vaginal, skin, and nasal) were selected for the kickoff of HMP, but the urinary tract was not selected because it was considered sterile. Historically, the sterility of the bladder/urine was established primarily based on bacterial-dependent detection methods. Before the clinical application of culture-independent molecular sequencing, it was speculated that healthy human urine was sterile, and this may have influenced standard microbial culture techniques [23]. Several recent studies have shown evidence of bacteria in the bladder of adult men and women without clinical infection (Table 1, Supplementary Table 1) [24-29].
The 16S ribosomal RNA (rRNA) gene sequencing and extended quantitative urine culture (EQUC) are techniques used primarily to detect bacterial DNA and live bacteria in cultured negative urine samples. The demonstration of microbiota in urine disproved the clinical dogma that urine is sterile. The association between the urinary microbiota and clinical factors rely on proper sampling of specific anatomical sites, and most studies in men are based on voided urine (Table 1). However, voided urine in patients does not truly represent the bladder microbiota, as the bacterial DNA detected in midstream void often differed substantially from the DNA detected in urine after catheterization. A study compared paired voided and catheterized urine obtained from men and found that the paired samples often did not match, providing evidence that voided urine does not truly characterize the bladder microbiota in men [30]. The lack of previous information on urinary tract microbiota in healthy men makes interpretation in clinical practice difficult. Therefore, standardized methodologies, including sampling should be developed. The early studies attempting to associate urologic diseases with urinary microbiota mainly focused on women [31-36]. In contrast to studies in men, studies in several women included a catheterized urine sample. Some Lactobacillus species are even associated with the absence of symptoms and defense against UTI [31,33]. These results support the notion that microbes can have protective abilities, and disruption of this microbial community may lead to urinary tract disorders. Some urinary tract diseases caused by microbial dysbiosis provide clues to research for the prevention, diagnosis, and treatment. Urine has been considered sterile for many years. However, urine is not sterile, and new knowledge related to urinary microbiota opens the door to a better understanding of lower urinary tract symptoms (LUTS). In women, compelling evidence associates lower urinary tract microbiota with LUTS [24]. Changes to low diversity urine microbiota dysbiosis were related with urge urinary incontinence (UUI) and interstitial cystitis/bladder pain syndrome (IC/BPS) [34,37,38].
Occult and recurrent cystitis may be due to invasion of the bladder wall by uropathogenic Escherichia coli (UPEC) and the formation of biofilm-like intracellular bacterial communities (IBCs). A possible explanation for LUTS is that IBCs formed after a UTI remain undetected by urine culture, evade antibiotic therapy, and act as a source of recurrent UTI (e.g., overactive bladder [OAB], IC/BPS) [39-41]. However, a similar relationship in men with benign prostate enlargement (BPE) has not been established. Most human studies have focused exclusively on bacteria, and very few investigations on fungi and viruses have been carried out [42-44]. It is necessary to reveal the contribution of other microbial communities such as fungi and viruses to diseases of the genitourinary system.
THE MALE URINARY MICROBIOTA IN BENIGN PROSTATE HYPERPLASIA AND LOWER URINARY TRACT SYMPTOMS
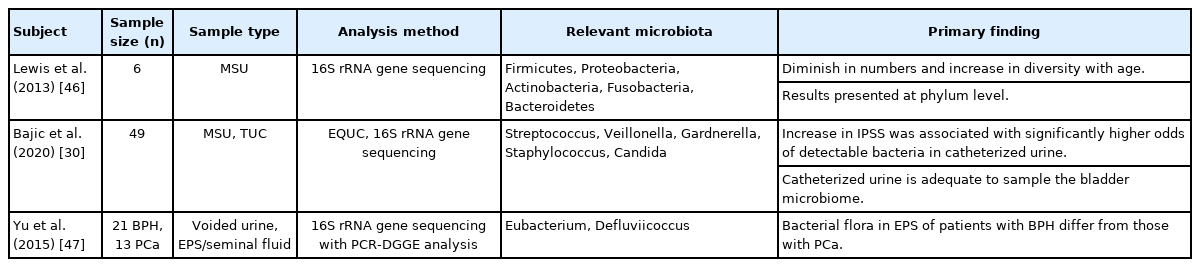
Although the female urinary microbiota has been well studied, there are only a few reports regarding the urinary microbiome in men (Table 2) [30, 45-47].
An early effort to identify the urinary microbiome in men was performed by Nelson et al. [25], by comparing men with sexually transmitted infection (STI) and STI-negative individuals. Lewis et al. [46] examined the effect of aging on the microbiota of male midstream voided urine and observed that the microbiota of the urethra appeared to be small in number and high in diversity as one grows old. But there is no study of the effects of aging on the microbiota of urine obtained by catheterization in men.
According to a study in women, the urinary microbiota may play a role in the pathogenesis of OAB, although the mechanisms underlying the causative relationship, as well as its possible therapeutic implications, are still uncertain [24]. The diversity of the urinary microbiome in patients with UUI was investigated by analyzing catheterized urine and was found to be decreased [34,38]. Therefore, urinary microbiome diversity in the urinary system of men should be evaluated for possible clinical application to LUTS in men.
BPH is a common urological disease that affects elderly men, causing LUTS. These symptoms can be divided into storage, voiding, and postmicturition symptoms [48]. LUTS are related to bladder outlet obstruction (BOO) caused by BPE due to histological changes in the prostate [48,49]. The α-blocker and 5-α reductase inhibitor have a therapeutic effect on symptoms of urination; however, eventually, 12.6% of patients suffer from worsening symptoms and approximately 5% of patients need surgical treatment [50,51]. These results support the need for more research on the physiological mechanisms and heterogeneity of LUTS in men [52]. Prostatic inflammation is known to play a key role in the pathogenesis and progression of BPH, but its causes are not well understood [19,20,53]. One of the possible causes is that the inhabitant bacterial microbiota of the lower urinary tract affects the local immunological environment [21]. The current pathophysiology of LUTS induced by BPE was based on the existing belief that the bladder is sterile. However, this belief was reconsidered after the presence of bacteria was demonstrated in the urine obtained from the bladder of men without a urinary tract infection (UTI). Further investigation was needed to evaluate the relationship between the male lower urinary microbiota and LUTS. As a clue to this, inflammatory cell infiltration of the prostate is associated with most patients with BPH and LUTS [54]. In the MTOPS (Medical Therapy of Prostatic Symptoms) study, 1,197 patients were biopsied, and histological evidence of prostate inflammation was present in 544 patients [54]. Patients with prostatic inflammation had significantly more acute urinary retention. It is possible to speculate the connection between tissue proliferation causing obstruction and inflammation. Prostatic inflammatory condition can stimulate interleukin (IL)-6 and IL-8 production. IL stimulates transforming growth factor synthesis and androgen receptors independently from dihydrotestosterone. Thus, in the state of prostatic inflammation, tissue proliferation may persist even in the presence of 5-α reductase inhibitors. In addition, proliferation and hypoxia stimulate the creation of reactive oxygen species [19]. This understanding of the inflammatory process of prostatic hyperplasia opens the avenue to explore new therapeutic possibilities.
Bajic et al. [30] analyzed microbes in urine between a group with 28 LUTS symptoms and undergoing surgery for BPE and a group with 21 LUTS symptoms not undergoing surgery. Using 16S rRNA sequencing and EQUC, they found the presence of dissimilar microbiota in the urethra and provided evidence of microbes in the bladder of male. Besides, in the catheterized urine of patients with high International Prostate Symptom Score (IPSS), the number of bacteria detected was higher (odds ratios, 2.21; P=0.029). Although there were no single species associated with LUTS in men, this study was the first to demonstrate the association of LUTS with the microbiome of the urinary system. This study showed that catheterized urine was more appropriate than voided urine for microbiota analysis. Using 16S rRNA sequencing, Yu et al. [47] compared the microbes of expressed-prostatic secretions (EPS), seminal fluid, and voided urine in patients with BPH and prostate cancer by and found that the microbiota of EPS was different in the 2 groups. BPH patients had higher numbers of species of the genera Eubacterium and Defluviicoccus, but lower numbers of the phyla Bacteroidetes, Alphaproteobacteria, and Firmicutes, as well as the family Lachnospiraceae and the genera Propionicimonas, Sphingomonas, and Ochrobactrum [47]. The genera Eubacterium and Defluviicoccus were founded more frequently in BPH patients but the phyla Bacteroidetes, Alphaproteobacteria, and Firmicutes were lower than in cancer patients [47]. The presence of certain bacteria can be caused by chronic inflammation of the prostate, which leads to an increase in proinflammatory cytokines. These studies suggest that ecological dysbiosis of the microbiota has a profound effect on prostate disease.
Bladder dysfunction may be included as one of the important causes of LUTS as well as diseases of the prostate [55]. Irritative voiding symptoms that occur in conditions such as UTI and BOO are triggered by inflammation. There is a possible role for inflammasomes, a cytosolic multiprotein oligomer of the innate immune system involved in activation of the inflammatory response, in the inflammation caused by bladder microbiota and prostate microbiota [56]. By understanding the role of each inflammasome in various pathological conditions, we may be able to target them with therapeutic agents to prevent these diseases and their symptoms. Some studies demonstrated the potential role of inflammasomes in changes in the bladder such as fibrosis and denervation [57-60]. The inflammasome assembled by nucleotide-binding domains, leucine-rich-containing families, and pyrin domain-containing-3, is simply known as the NLRP3 inflammasome, which is present in cells that recognize pathogen-related molecular patterns and damage-associated molecular patterns (DAMPs) [61]. Inflammasome-activating insults caused by BOO are DAMPs by hypoxia/reperfusion, increased pressure, and repetitive stretching [56]. BOO triggers a progressive inflammatory process mediated by NLRP3 that triggers negative downstream events such as irritative voiding and fibrosis. Inflammasomes may mediate both infectious and sterile prostatitis, leading to BPH, BOO, and activation of NLRP3 within the bladder, eventually causing all the harmful effects such as irritative voiding, bladder fibrosis, and denervation. The main NLR associated with the prostate is thought to be NLRP1, not NLRP3, and NLRP1 plays an important role in prostate inflammation [62,63]. It differs mechanistically from NLRP3 because it undergoes autolytic proteolysis necessary for activation [64].
CHRONIC PROSTATITIS/CHRONIC PELVIC PAIN SYNDROME AND THE URINARY MICROBIOTA
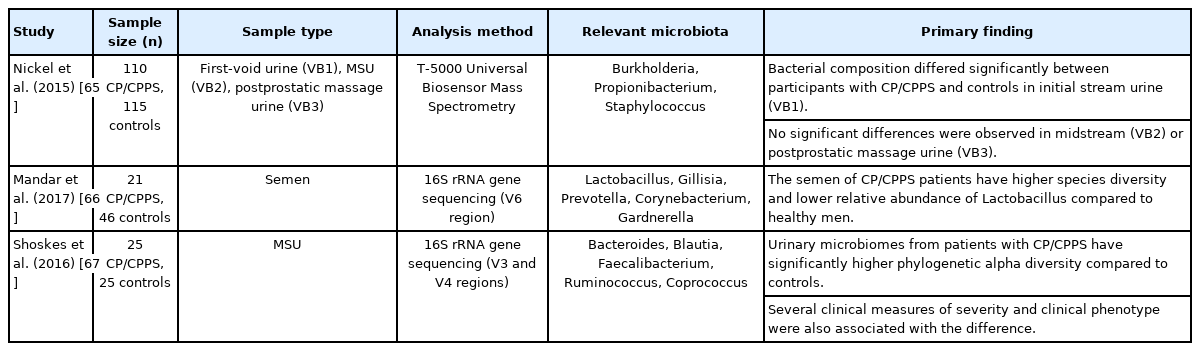
Several researches have investigated the role of the urinary microbiota in CP/CPPS (Table 3) [65-67].
Nickel et al. examined first-void (VB1), midstream void (VB2), and postprostatic massage void (VB3) urines from 110 CP/CPPS patients and 115 controls [68] in the multidisciplinary [65]. Overall species and genus composition differed significantly for first-void urine only (VB1). The bacterial species Burkholderia cenocepacia was confirmed to be overexpressed in the CP/CPPS population [65]. They did not find any putative organisms for CP/CPPS, but the specific microbiome differences observed for B. cenocepacia may indicate a change in overall species balance. Other researchers described B. cenocepacia as a pathogen, presuming that it was involved in the pathogenesis of CP/CPPS [69-71].
Shoskes et al. [67] compared 25 patients with CP/CPPS and 25 controls using 16S rRNA gene sequencing. In the CP/CPPS patients, the phylogenetic diversity was confirmed by the overexpression of 17 bacterial taxa, and they also had more Clostridia and Bacterodia bacterial species. It is unclear why the phylogenetic diversity of the urine microbiota is greater in CP/CPPS patients who are more frequently used antibiotics. Patients have a high prevalence of anaerobic bacteria, which indicates pathogens that are not usually cultured or treated in clinical practice
Shoskes et al. [72] recently investigated the role of intestinal microbiomes in CP/CPPS. It is believed that intestinal microbiomes can affect the symptoms or clinical phenotype of CP/CPPS patients. In this study, Prevotella was found to be dominant in the intestines of the control group, which can be assumed to optimize energy intake and prevent inflammation. Therefore, having less Prevotella in the CP/CPPS patient’s intestine could be considered as one of the etiologies. The well-balanced microbiota is in a state of “eubiosis” that functions smoothly for the whole organism. Meanwhile, qualitative and quantitative changes of microbiota are called “dysbiosis.” Thus, treatment approaches could aim to restore the microbiota by removing bacteria and growth and inflammation-causing toxins produced by the microorganism [73].
Mandar et al. [66] studied the semen of 21 men with CP/CPPS and 46 controls using 16S rRNA gene sequencing. They showed that the difference between these 2 groups is relative depletion in the genus Lactobacillus. In patients with prostatitis, the relative abundance of the species Lactobacillus iners was significantly lower. In addition, they noted greater microbial diversity in patients with prostatitis.
Murphy et al. [74,75] isolated certain Staphylococcus epidermal strains from expressed prostate secretions from healthy human males and performed intraurethral instillation using murine experimental prostatitis. They reported that the instillation reduced the pelvic tactile allodynia responses and the increased T-cell numbers associated with prostatitis. Their results showed new possibilities for commensal Staphylococcus epidermis and its cellular components in the treatment of prostatitis-related pain.
Because the pathophysiology of CP/CPPS is not well understood, the research of the urogenital microbiota could be used not only to understand the pathology of the disease but also to explore treatments that can restore eubiosis and prevent the vicious cycle of dysbiosis-urogenital infections.
CONCLUSION
The urinary tract, once believed to be sterile, has now been shown to harbor bacteria. Since then, new approaches to studying the disorders of the urinary tract have emerged. We are beginning to understand the potential role of microbial communities in the urinary tract in the pathogenesis of urogenital diseases, and some preliminary results suggest that the microbiota may play a role in a variety of ways. However, the evidence is still not compelling and, therefore, difficult to use in clinical practice. Besides, the fact that microbiome-related research is not unified, it poses an obstacle to the interpretation of studies. Also, as new understandings of the inflammatory response and inflammasome in diseases of the urinary tract are emerging, the process of finding the link between the microbiome and inflammatory response is expected to play an important role in advancing the approach to interrogate and manage diseases of the urinary system.
Prostatic disease should continue to be reevaluated and diseases (BPH/LUTS, CP/CPPS) previously thought to be unrelated to microbes should be thoroughly investigated. Perhaps with more study, we can develop the means to handle the urinary microbiome to improve patient outcomes.
SUPPLEMENTARY MATERIALS
Supplementary Table 1.
Notes
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
AUTHOR CONTRIBUTION STATEMENT
·Conceptualization: SIJ
·Data curation: SIJ
·Writing-original draft: MSK, SIJ
·Writing-review & editing: MSK, SIJ